Promoting developmental transcription
- PMID: 20023156
- PMCID: PMC2796937
- DOI: 10.1242/dev.035493
Promoting developmental transcription
Abstract
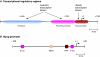
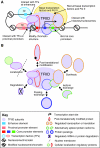
Animal growth and development depend on the precise control of gene expression at the level of transcription. A central role in the regulation of developmental transcription is attributed to transcription factors that bind DNA enhancer elements, which are often located far from gene transcription start sites. Here, we review recent studies that have uncovered significant regulatory functions in developmental transcription for the TFIID basal transcription factors and for the DNA core promoter elements that are located close to transcription start sites.
Figures



Similar articles
-
Novel cofactors and TFIIA mediate functional core promoter selectivity by the human TAFII150-containing TFIID complex.Mol Cell Biol. 1998 Nov;18(11):6571-83. doi: 10.1128/MCB.18.11.6571. Mol Cell Biol. 1998. PMID: 9774672 Free PMC article.
-
Taspase1 processing alters TFIIA cofactor properties in the regulation of TFIID.Transcription. 2015;6(2):21-32. doi: 10.1080/21541264.2015.1052178. Transcription. 2015. PMID: 25996597 Free PMC article.
-
RNA polymerase II and TAFs undergo a slow isomerization after the polymerase is recruited to promoter-bound TFIID.J Mol Biol. 2010 Mar 19;397(1):57-68. doi: 10.1016/j.jmb.2010.01.025. Epub 2010 Jan 18. J Mol Biol. 2010. PMID: 20083121
-
Shifting players and paradigms in cell-specific transcription.Mol Cell. 2009 Dec 25;36(6):924-31. doi: 10.1016/j.molcel.2009.12.011. Mol Cell. 2009. PMID: 20064459 Free PMC article. Review.
-
An unexpected role of TAFs and TRFs in skeletal muscle differentiation: switching core promoter complexes.Cold Spring Harb Symp Quant Biol. 2008;73:217-25. doi: 10.1101/sqb.2008.73.028. Epub 2008 Nov 6. Cold Spring Harb Symp Quant Biol. 2008. PMID: 19022758 Review.
Cited by
-
Highly redundant function of multiple AT-rich sequences as core promoter elements in the TATA-less RPS5 promoter of Saccharomyces cerevisiae.Nucleic Acids Res. 2011 Jan;39(1):59-75. doi: 10.1093/nar/gkq741. Epub 2010 Aug 30. Nucleic Acids Res. 2011. PMID: 20805245 Free PMC article.
-
Chromatin-associated SUMOylation controls the transcriptional switch between plant development and heat stress responses.Plant Commun. 2020 Jul 2;2(1):100091. doi: 10.1016/j.xplc.2020.100091. eCollection 2021 Jan 11. Plant Commun. 2020. PMID: 33511343 Free PMC article.
-
The Core Promoter Is a Regulatory Hub for Developmental Gene Expression.Front Cell Dev Biol. 2021 Sep 10;9:666508. doi: 10.3389/fcell.2021.666508. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34568311 Free PMC article. Review.
-
A Random Screen Using a Novel Reporter Assay System Reveals a Set of Sequences That Are Preferred as the TATA or TATA-Like Elements in the CYC1 Promoter of Saccharomyces cerevisiae.PLoS One. 2015 Jun 5;10(6):e0129357. doi: 10.1371/journal.pone.0129357. eCollection 2015. PLoS One. 2015. PMID: 26046838 Free PMC article.
-
Incorporating Sequence-Dependent DNA Shape and Dynamics into Transcriptome Data Analysis.Methods Mol Biol. 2024;2812:317-343. doi: 10.1007/978-1-0716-3886-6_18. Methods Mol Biol. 2024. PMID: 39068371
References
-
- Auty R., Steen H., Myers L. C., Persinger J., Bartholomew B., Gygi S. P., Buratowski S. (2004). Purification of active TFIID from Saccharomyces cerevisiae: Extensive promoter contact and co-activator function. J. Biol. Chem. 279, 49973-49981 - PubMed
-
- Bártfai R., Balduf C., Hilton T., Rathmann Y., Hadzhiev Y., Tora L., Orbán L., Müller F. (2004). TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr. Biol. 14, 593-598 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

