FGF-receptor signalling controls neural cell diversity in the zebrafish hindbrain by regulating olig2 and sox9
- PMID: 20023158
- PMCID: PMC2796930
- DOI: 10.1242/dev.038026
FGF-receptor signalling controls neural cell diversity in the zebrafish hindbrain by regulating olig2 and sox9
Abstract
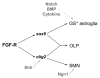
The mechanisms underlying the generation of neural cell diversity are the subject of intense investigation, which has highlighted the involvement of different signalling molecules including Shh, BMP and Wnt. By contrast, relatively little is known about FGF in this process. In this report we identify an FGF-receptor-dependent pathway in zebrafish hindbrain neural progenitors that give rise to somatic motoneurons, oligodendrocyte progenitors and differentiating astroglia. Using a combination of chemical and genetic approaches to conditionally inactivate FGF-receptor signalling, we investigate the role of this pathway. We show that FGF-receptor signalling is not essential for the survival or maintenance of hindbrain neural progenitors but controls their fate by coordinately regulating key transcription factors. First, by cooperating with Shh, FGF-receptor signalling controls the expression of olig2, a patterning gene essential for the specification of somatic motoneurons and oligodendrocytes. Second, FGF-receptor signalling controls the development of both oligodendrocyte progenitors and astroglia through the regulation of sox9, a gliogenic transcription factor the function of which we show to be conserved in the zebrafish hindbrain. Overall, for the first time in vivo, our results reveal a mechanism of FGF in the control of neural cell diversity.
Figures






Similar articles
-
Short-range Fgf signalling patterns hindbrain progenitors to induce the neurogenesis-to-oligodendrogenesis switch.Development. 2024 Dec 15;151(24):dev204256. doi: 10.1242/dev.204256. Epub 2024 Dec 13. Development. 2024. PMID: 39575980 Free PMC article.
-
Olig2+ precursors produce abducens motor neurons and oligodendrocytes in the zebrafish hindbrain.J Neurosci. 2009 Feb 25;29(8):2322-33. doi: 10.1523/JNEUROSCI.3755-08.2009. J Neurosci. 2009. PMID: 19244509 Free PMC article.
-
Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS.Mech Dev. 2006 Jan;123(1):24-30. doi: 10.1016/j.mod.2005.10.005. Epub 2005 Dec 1. Mech Dev. 2006. PMID: 16324829
-
Autotaxin/ENPP2 regulates oligodendrocyte differentiation in vivo in the developing zebrafish hindbrain.Glia. 2012 Oct;60(10):1605-18. doi: 10.1002/glia.22381. Epub 2012 Jul 20. Glia. 2012. PMID: 22821873 Free PMC article.
-
Transport and gradient formation of Wnt and Fgf in the early zebrafish gastrula.Curr Top Dev Biol. 2024;157:125-153. doi: 10.1016/bs.ctdb.2023.12.003. Epub 2023 Dec 27. Curr Top Dev Biol. 2024. PMID: 38556457 Review.
Cited by
-
Hindbrain rhombomere centers harbor a heterogenous population of dividing progenitors which rely on Notch signaling.Front Cell Dev Biol. 2023 Nov 2;11:1268631. doi: 10.3389/fcell.2023.1268631. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 38020924 Free PMC article.
-
Peptide ligands targeting FGF receptors promote recovery from dorsal root crush injury via AKT/mTOR signaling.Theranostics. 2021 Nov 2;11(20):10125-10147. doi: 10.7150/thno.62525. eCollection 2021. Theranostics. 2021. PMID: 34815808 Free PMC article.
-
FGF signaling controls Shh-dependent oligodendroglial fate specification in the ventral spinal cord.Neural Dev. 2018 Mar 8;13(1):3. doi: 10.1186/s13064-018-0100-2. Neural Dev. 2018. PMID: 29519242 Free PMC article.
-
Brorin is required for neurogenesis, gliogenesis, and commissural axon guidance in the zebrafish forebrain.PLoS One. 2017 Apr 27;12(4):e0176036. doi: 10.1371/journal.pone.0176036. eCollection 2017. PLoS One. 2017. PMID: 28448525 Free PMC article.
-
Nucleolin loss of function leads to aberrant Fibroblast Growth Factor signaling and craniofacial anomalies.Development. 2022 Jun 15;149(12):dev200349. doi: 10.1242/dev.200349. Epub 2022 Jun 28. Development. 2022. PMID: 35762670 Free PMC article.
References
-
- Amoyel M., Cheng Y. C., Jiang Y. J., Wilkinson D. G. (2005). Wnt1 regulates neurogenesis and mediates lateral inhibition of boundary cell specification in the zebrafish hindbrain. Development 132, 775-785 - PubMed
-
- Bertrand N., Castro D. S., Guillemot F. (2002). Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517-530 - PubMed
-
- Briscoe J., Ericson J. (1999). The specification of neuronal identity by graded Sonic Hedgehog signalling. Semin. Cell Dev. Biol. 10, 353-362 - PubMed
-
- Cai J., Qi Y., Hu X., Tan M., Liu Z., Zhang J., Li Q., Sander M., Qiu M. (2005). Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron 45, 41-53 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

