Control of cell proliferation in Arabidopsis thaliana by microRNA miR396
- PMID: 20023165
- PMCID: PMC2796936
- DOI: 10.1242/dev.043067
Control of cell proliferation in Arabidopsis thaliana by microRNA miR396
Abstract
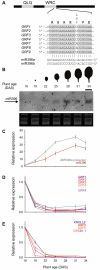
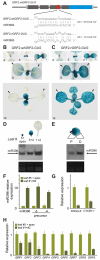
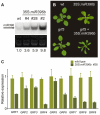
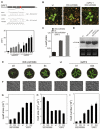
Cell proliferation is an important determinant of plant form, but little is known about how developmental programs control cell division. Here, we describe the role of microRNA miR396 in the coordination of cell proliferation in Arabidopsis leaves. In leaf primordia, miR396 is expressed at low levels that steadily increase during organ development. We found that miR396 antagonizes the expression pattern of its targets, the GROWTH-REGULATING FACTOR (GRF) transcription factors. miR396 accumulates preferentially in the distal part of young developing leaves, restricting the expression of GRF2 to the proximal part of the organ. This, in turn, coincides with the activity of the cell proliferation marker CYCLINB1;1. We show that miR396 attenuates cell proliferation in developing leaves, through the repression of GRF activity and a decrease in the expression of cell cycle genes. We observed that the balance between miR396 and the GRFs controls the final number of cells in leaves. Furthermore, overexpression of miR396 in a mutant lacking GRF-INTERACTING FACTOR 1 severely compromises the shoot meristem. We found that miR396 is expressed at low levels throughout the meristem, overlapping with the expression of its target, GRF2. In addition, we show that miR396 can regulate cell proliferation and the size of the meristem. Arabidopsis plants with an increased activity of the transcription factor TCP4, which reduces cell proliferation in leaves, have higher miR396 and lower GRF levels. These results implicate miR396 as a significant module in the regulation of cell proliferation in plants.
Figures


Similar articles
-
Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions.PLoS Genet. 2012 Jan;8(1):e1002419. doi: 10.1371/journal.pgen.1002419. Epub 2012 Jan 5. PLoS Genet. 2012. PMID: 22242012 Free PMC article.
-
Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis.Physiol Plant. 2009 Jun;136(2):223-36. doi: 10.1111/j.1399-3054.2009.01229.x. Epub 2009 Feb 2. Physiol Plant. 2009. PMID: 19453503
-
MicroRNA miR396 and RDR6 synergistically regulate leaf development.Mech Dev. 2013 Jan;130(1):2-13. doi: 10.1016/j.mod.2012.07.005. Epub 2012 Aug 4. Mech Dev. 2013. PMID: 22889666
-
Growth-regulating factors: conserved and divergent roles in plant growth and development and potential value for crop improvement.Plant J. 2023 Mar;113(6):1122-1145. doi: 10.1111/tpj.16090. Epub 2023 Jan 23. Plant J. 2023. PMID: 36582168
-
Control of cell proliferation by microRNAs in plants.Curr Opin Plant Biol. 2016 Dec;34:68-76. doi: 10.1016/j.pbi.2016.10.003. Epub 2016 Oct 26. Curr Opin Plant Biol. 2016. PMID: 27794260 Review.
Cited by
-
Identification and characterization of microRNAs from barley (Hordeum vulgare L.) by high-throughput sequencing.Int J Mol Sci. 2012;13(3):2973-2984. doi: 10.3390/ijms13032973. Epub 2012 Mar 6. Int J Mol Sci. 2012. PMID: 22489137 Free PMC article.
-
Expression-based functional investigation of the organ-specific microRNAs in Arabidopsis.PLoS One. 2012;7(11):e50870. doi: 10.1371/journal.pone.0050870. Epub 2012 Nov 30. PLoS One. 2012. PMID: 23226412 Free PMC article.
-
Genome-Wide Identification and Characterization of Growth Regulatory Factor Family Genes in Medicago.Int J Mol Sci. 2022 Jun 21;23(13):6905. doi: 10.3390/ijms23136905. Int J Mol Sci. 2022. PMID: 35805911 Free PMC article.
-
Coordination between GROWTH-REGULATING FACTOR1 and GRF-INTERACTING FACTOR1 plays a key role in regulating leaf growth in rice.BMC Plant Biol. 2020 May 8;20(1):200. doi: 10.1186/s12870-020-02417-0. BMC Plant Biol. 2020. PMID: 32384927 Free PMC article.
-
Identification and comparative analysis of cadmium tolerance-associated miRNAs and their targets in two soybean genotypes.PLoS One. 2013 Dec 10;8(12):e81471. doi: 10.1371/journal.pone.0081471. eCollection 2013. PLoS One. 2013. PMID: 24363811 Free PMC article.
References
-
- Baker C. C., Sieber P., Wellmer F., Meyerowitz E. M. (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 15, 303-315 - PubMed
-
- Bartel D. P., Chen C. Z. (2004). Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5, 396-400 - PubMed
-
- Cartolano M., Castillo R., Efremova N., Kuckenberg M., Zethof J., Gerats T., Schwarz-Sommer Z., Vandenbussche M. (2007). A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat. Genet. 39, 901-905 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

