Activation of the chloroplast monogalactosyldiacylglycerol synthase MGD1 by phosphatidic acid and phosphatidylglycerol
- PMID: 20023301
- PMCID: PMC2825394
- DOI: 10.1074/jbc.M109.071928
Activation of the chloroplast monogalactosyldiacylglycerol synthase MGD1 by phosphatidic acid and phosphatidylglycerol
Abstract
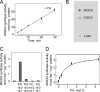
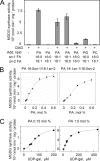
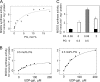
One of the major characteristics of chloroplast membranes is their enrichment in galactoglycerolipids, monogalactosyldiacylglycerol (MGDG), and digalactosyldiacylglycerol (DGDG), whereas phospholipids are poorly represented, mainly as phosphatidylglycerol (PG). All these lipids are synthesized in the chloroplast envelope, but galactolipid synthesis is also partially dependent on phospholipid synthesis localized in non-plastidial membranes. MGDG synthesis was previously shown essential for chloroplast development. In this report, we analyze the regulation of MGDG synthesis by phosphatidic acid (PA), which is a general precursor in the synthesis of all glycerolipids and is also a signaling molecule in plants. We demonstrate that under physiological conditions, MGDG synthesis is not active when the MGDG synthase enzyme is supplied with its substrates only, i.e. diacylglycerol and UDP-gal. In contrast, PA activates the enzyme when supplied. This is shown in leaf homogenates, in the chloroplast envelope, as well as on the recombinant MGDG synthase, MGD1. PG can also activate the enzyme, but comparison of PA and PG effects on MGD1 activity indicates that PA and PG proceed through different mechanisms, which are further differentiated by enzymatic analysis of point-mutated recombinant MGD1s. Activation of MGD1 by PA and PG is proposed as an important mechanism coupling phospholipid and galactolipid syntheses in plants.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

