Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats
- PMID: 20023628
- PMCID: PMC2811870
- DOI: 10.1038/nature08648
Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats
Abstract
Broken chromosomes arising from DNA double-strand breaks result from endogenous events such as the production of reactive oxygen species during cellular metabolism, as well as from exogenous sources such as ionizing radiation. Left unrepaired or incorrectly repaired they can lead to genomic changes that may result in cell death or cancer. DNA-dependent protein kinase (DNA-PK), a holoenzyme that comprises the DNA-PK catalytic subunit (DNA-PKcs) and the heterodimer Ku70/Ku80, has a major role in non-homologous end joining-the main pathway in mammals used to repair double-strand breaks. DNA-PKcs is a serine/threonine protein kinase comprising a single polypeptide chain of 4,128 amino acids and belonging to the phosphatidylinositol-3-OH kinase (PI(3)K)-related protein family. DNA-PKcs is involved in the sensing and transmission of DNA damage signals to proteins such as p53, setting off events that lead to cell cycle arrest. It phosphorylates a wide range of substrates in vitro, including Ku70/Ku80, which is translocated along DNA. Here we present the crystal structure of human DNA-PKcs at 6.6 A resolution, in which the overall fold is clearly visible, to our knowledge, for the first time. The many alpha-helical HEAT repeats (helix-turn-helix motifs) facilitate bending and allow the polypeptide chain to fold into a hollow circular structure. The carboxy-terminal kinase domain is located on top of this structure, and a small HEAT repeat domain that probably binds DNA is inside. The structure provides a flexible cradle to promote DNA double-strand-break repair.
Figures
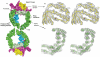
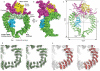

Similar articles
-
Cryo-EM structure of human DNA-PK holoenzyme.Cell Res. 2017 Nov;27(11):1341-1350. doi: 10.1038/cr.2017.110. Epub 2017 Aug 25. Cell Res. 2017. PMID: 28840859 Free PMC article.
-
The N-terminal region of the DNA-dependent protein kinase catalytic subunit is required for its DNA double-stranded break-mediated activation.J Biol Chem. 2013 Mar 8;288(10):7037-46. doi: 10.1074/jbc.M112.434498. Epub 2013 Jan 15. J Biol Chem. 2013. PMID: 23322783 Free PMC article.
-
Cryo-EM structure of the DNA-PK holoenzyme.Proc Natl Acad Sci U S A. 2017 Jul 11;114(28):7367-7372. doi: 10.1073/pnas.1707386114. Epub 2017 Jun 26. Proc Natl Acad Sci U S A. 2017. PMID: 28652322 Free PMC article.
-
DNA-Dependent Protein Kinase Catalytic Subunit: The Sensor for DNA Double-Strand Breaks Structurally and Functionally Related to Ataxia Telangiectasia Mutated.Genes (Basel). 2021 Jul 27;12(8):1143. doi: 10.3390/genes12081143. Genes (Basel). 2021. PMID: 34440313 Free PMC article. Review.
-
One ring to bring them all--the role of Ku in mammalian non-homologous end joining.DNA Repair (Amst). 2014 May;17:30-8. doi: 10.1016/j.dnarep.2014.02.019. Epub 2014 Mar 26. DNA Repair (Amst). 2014. PMID: 24680220 Review.
Cited by
-
ATR-mediated regulation of nuclear and cellular plasticity.DNA Repair (Amst). 2016 Aug;44:143-150. doi: 10.1016/j.dnarep.2016.05.020. Epub 2016 May 16. DNA Repair (Amst). 2016. PMID: 27283761 Free PMC article. Review.
-
Structure of the human dimeric ATM kinase.Cell Cycle. 2016;15(8):1117-24. doi: 10.1080/15384101.2016.1158362. Cell Cycle. 2016. PMID: 27097373 Free PMC article.
-
Chemotherapeutic compounds targeting the DNA double-strand break repair pathways: the good, the bad, and the promising.Front Oncol. 2014 Apr 22;4:86. doi: 10.3389/fonc.2014.00086. eCollection 2014. Front Oncol. 2014. PMID: 24795863 Free PMC article. Review.
-
Mutation of serine 1333 in the ATR HEAT repeats creates a hyperactive kinase.PLoS One. 2014 Jun 5;9(6):e99397. doi: 10.1371/journal.pone.0099397. eCollection 2014. PLoS One. 2014. PMID: 24901225 Free PMC article.
-
Interaction between HIV-1 Tat and DNA-PKcs modulates HIV transcription and class switch recombination.Int J Biol Sci. 2014 Oct 9;10(10):1138-49. doi: 10.7150/ijbs.10366. eCollection 2014. Int J Biol Sci. 2014. PMID: 25332688 Free PMC article.
References
-
- Kemp LM, Sedgwick SG, Jeggo PA. X-ray sensitive mutants of Chinese hamster ovary cells defective in double-strand break rejoining. Mutat. Res. 1984;132:189–196. - PubMed
-
- Zdzienicka MZ, Tran Q, van der Schans GP, Simons JWI. Characterization of an X-ray-hypersensitive mutant of V79 Chinese hamster cells. Mutat. Res. 1988;194:239–249. - PubMed
-
- Dvir A, Stein LY, Calore BL, Dynan WS. Purification and characterization of a template associated protein kinase that phosphorylates RNA polymerase II. J. Biol. Chem. 1993;268:10440–10447. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

