Microparticle encoding technologies for high-throughput multiplexed suspension assays
- PMID: 20023742
- PMCID: PMC7108550
- DOI: 10.1039/b905502a
Microparticle encoding technologies for high-throughput multiplexed suspension assays
Abstract
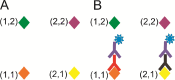
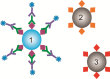
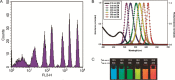
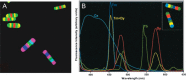
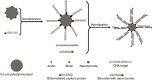
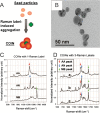
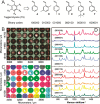
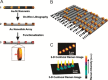
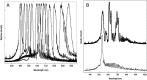
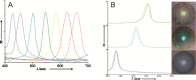
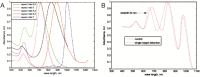
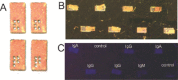
The requirement for analysis of large numbers of biomolecules for drug discovery and clinical diagnostics has driven the development of low-cost, flexible and high-throughput methods for simultaneous detection of multiple molecular targets in a single sample (multiplexed analysis). The technique that seems most likely to satisfy all of these requirements is the multiplexed suspension (bead-based) assay, which offers a number of advantages over alternative approaches such as ELISAs and microarrays. In a bead based assay, different probe molecules are attached to different beads (of a few tens of microns in size), which are then reacted in suspension with the target sample. After reaction, the beads must be identifiable in order to determine the attached probe molecule, and thus each bead must be labelled (encoded) with a unique identifier. A large number of techniques have been proposed for encoding beads. This critical review analyses each technology on the basis of its ability to fulfil the practical requirements of assays, whilst being compatible with low-cost, high-throughput manufacturing processes and high-throughput detection methods. As a result, we identify the most likely candidates to be used for future integrated device development for practical applications.
Figures




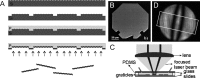
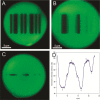



References
-
- Kindt T. J., Goldsby R. A. andOsborne B. A.,Immunology,W. H. Freeman and Company,New York, 6th edn,2007.
-
- AbCam ELISA Kits specifications: http://www.abcam.com.
-
- Quansys Bio multiplex ELISA kit specifications: http://www.quansysbio.com/.
-
- Pierce multiplex ELISA kit specifications: http://www.piercenet.com/.
-
- Ray Biotech multiplex ELISA kit specifications:http://www.raybiotech.com/.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

