Human mammary progenitor cell fate decisions are products of interactions with combinatorial microenvironments
- PMID: 20023793
- PMCID: PMC2933184
- DOI: 10.1039/b816472j
Human mammary progenitor cell fate decisions are products of interactions with combinatorial microenvironments
Abstract
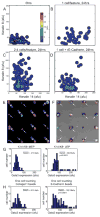
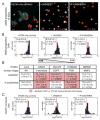
In adult tissues, multi-potent progenitor cells are some of the most primitive members of the developmental hierarchies that maintain homeostasis. That progenitors and their more mature progeny share identical genomes, suggests that fate decisions are directed by interactions with extrinsic soluble factors, ECM, and other cells, as well as physical properties of the ECM. To understand regulation of fate decisions, therefore, would require a means of understanding carefully choreographed combinatorial interactions. Here we used microenvironment protein microarrays to functionally identify combinations of cell-extrinsic mammary gland proteins and ECM molecules that imposed specific cell fates on bipotent human mammary progenitor cells. Micropatterned cell culture surfaces were fabricated to distinguish between the instructive effects of cell-cell versus cell-ECM interactions, as well as constellations of signaling molecules; and these were used in conjunction with physiologically relevant 3 dimensional human breast cultures. Both immortalized and primary human breast progenitors were analyzed. We report on the functional ability of those proteins of the mammary gland that maintain quiescence, maintain the progenitor state, and guide progenitor differentiation towards myoepithelial and luminal lineages.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

