Epigenome manipulation as a pathway to new natural product scaffolds and their congeners
- PMID: 20024091
- PMCID: PMC2958777
- DOI: 10.1039/b920860g
Epigenome manipulation as a pathway to new natural product scaffolds and their congeners
Abstract
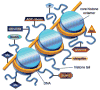
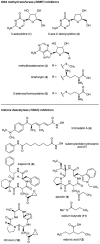
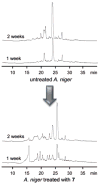
The covalent modification of chromatin is an important control mechanism used by fungi to modulate the transcription of genes involved in secondary metabolite production. To date, both molecular-based and chemical approaches targeting histone and DNA posttranslational processes have shown great potential for rationally directing the activation and/or suppression of natural-product-encoding gene clusters. In this Highlight, the organization of the fungal epigenome is summarized and strategies for manipulating chromatin-related targets are presented. Applications of these techniques are illustrated using several recently published accounts in which chemical-epigenetic methods and mutant studies were successfully employed for the de novo or enhanced production of structurally diverse fungal natural products (e.g., anthraquinones, cladochromes, lunalides, mycotoxins, and nygerones).
Figures



Similar articles
-
Fungal extrolites as a new source for therapeutic compounds and as building blocks for applications in synthetic biology.Microbiol Res. 2014 Sep-Oct;169(9-10):652-65. doi: 10.1016/j.micres.2014.02.007. Epub 2014 Feb 25. Microbiol Res. 2014. PMID: 24636745 Review.
-
Traversing the fungal terpenome.Nat Prod Rep. 2014 Oct;31(10):1449-73. doi: 10.1039/c4np00075g. Nat Prod Rep. 2014. PMID: 25171145 Free PMC article. Review.
-
Harnessing diverse transcriptional regulators for natural product discovery in fungi.Nat Prod Rep. 2020 Jan 29;37(1):6-16. doi: 10.1039/c8np00027a. Nat Prod Rep. 2020. PMID: 31033969 Review.
-
Small-molecule elicitation of microbial secondary metabolites.Microb Biotechnol. 2011 Jul;4(4):471-8. doi: 10.1111/j.1751-7915.2010.00196.x. Epub 2010 Jul 26. Microb Biotechnol. 2011. PMID: 21375710 Free PMC article. Review.
-
Diversifying microbial natural products for drug discovery.Appl Microbiol Biotechnol. 2003 Oct;62(5-6):446-58. doi: 10.1007/s00253-003-1381-9. Epub 2003 Jun 28. Appl Microbiol Biotechnol. 2003. PMID: 12838377 Review.
Cited by
-
Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites.PLoS Pathog. 2013;9(6):e1003475. doi: 10.1371/journal.ppat.1003475. Epub 2013 Jun 27. PLoS Pathog. 2013. PMID: 23825955 Free PMC article.
-
An epigenetic modifier induces production of 3-(4-oxopyrano)-chromen-2-ones in Aspergillus sp. AST0006, an endophytic fungus of Astragalus lentiginosus.Tetrahedron. 2020 Oct 23;76(43):131525. doi: 10.1016/j.tet.2020.131525. Epub 2020 Aug 22. Tetrahedron. 2020. PMID: 33716326 Free PMC article.
-
Reactivation of antibiosis in the entomogenous fungus Chrysoporthe sp. SNB-CN74.J Antibiot (Tokyo). 2015 Sep;68(9):586-90. doi: 10.1038/ja.2015.36. Epub 2015 Apr 15. J Antibiot (Tokyo). 2015. PMID: 25873318
-
Insights into microbial cryptic gene activation and strain improvement: principle, application and technical aspects.J Antibiot (Tokyo). 2017 Jan;70(1):25-40. doi: 10.1038/ja.2016.82. Epub 2016 Jul 6. J Antibiot (Tokyo). 2017. PMID: 27381522 Review.
-
Modulation of polyketide biosynthetic pathway of the endophytic fungus, Anteaglonium sp. FL0768, by copper (II) and anacardic acid.Phytochem Lett. 2018 Dec;28:157-163. doi: 10.1016/j.phytol.2018.10.011. Epub 2018 Oct 25. Phytochem Lett. 2018. PMID: 31354886 Free PMC article.
References
-
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D'Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Nature. 2005;438:1105–1115. - PubMed
-
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CPC, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, Birren B. Nature. 2003;422:859–868. - PubMed
-
- Fisch KM, Gillaspy AF, Gipson M, Henrikson JC, Hoover AR, Jackson L, Najar FZ, Wägele H, Cichewicz RH. J Ind Microbiol Biotechnol. 2009;36:1199–1213. - PubMed
-
- Corre C, Challis GL. Nat Prod Rep. 2009;26:977–986. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

