Improving limits of detection for B-type natriuretic peptide using PC-IDMS: an application of the ALiPHAT strategy
- PMID: 20024179
- PMCID: PMC3129710
- DOI: 10.1039/b919484c
Improving limits of detection for B-type natriuretic peptide using PC-IDMS: an application of the ALiPHAT strategy
Abstract
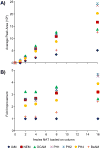
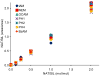
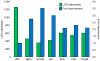
Hydrophobic tagging of biomolecules has been reported by our group and others to increase their ionization efficiency during electrospray ionization and facilitate their detection by mass spectrometry. As such, hydrophobic tagging should provide a viable method for augmenting MS-based quantification of low abundance proteins by decreasing their detection limits. Herein we have evaluated two commercial alkylation reagents and several newly synthesized hydrophobic alkylation reagents for their utility in quantifying B-type Natriuretic Peptide, a low abundance cardiac biomarker, by protein cleavage isotope dilution mass spectrometry. For the cysteine containing tryptic peptide evaluated, a approximately 3.5-fold decrease in the detection limit was observed for the best performing hydrophobic reagent, 2-iodo-N-octylacetamide, relative to the commonly used alkylation reagent, iodoacetamide. Additionally, we have evaluated the use of nonpolar surface areas as a metric for assessing the effectiveness of the alkylation reagents in improving ESI response.
Figures
Similar articles
-
Alkylating tryptic peptides to enhance electrospray ionization mass spectrometry analysis.Anal Chem. 2010 Dec 15;82(24):10135-42. doi: 10.1021/ac1019792. Epub 2010 Nov 29. Anal Chem. 2010. PMID: 21114270 Free PMC article.
-
Evaluation of the ALiPHAT method for PC-IDMS and correlation of limits-of-detection with nonpolar surface area.J Am Soc Mass Spectrom. 2009 Nov;20(11):2006-12. doi: 10.1016/j.jasms.2009.07.019. Epub 2009 Aug 7. J Am Soc Mass Spectrom. 2009. PMID: 19734056 Free PMC article.
-
N-t-butyliodoacetamide and iodoacetanilide: two new cysteine alkylating reagents for relative quantitation of proteins.Rapid Commun Mass Spectrom. 2004;18(1):117-27. doi: 10.1002/rcm.1286. Rapid Commun Mass Spectrom. 2004. PMID: 14689568
-
Molecules and elements for quantitative bioanalysis: The allure of using electrospray, MALDI, and ICP mass spectrometry side-by-side.Mass Spectrom Rev. 2019 Mar;38(2):169-186. doi: 10.1002/mas.21567. Epub 2018 Mar 30. Mass Spectrom Rev. 2019. PMID: 29603315 Review.
-
Derivatization in liquid chromatography for mass spectrometric detection.Drug Discov Ther. 2013 Feb;7(1):9-17. doi: 10.5582/ddt.2013.v7.1.9. Drug Discov Ther. 2013. PMID: 23524938 Review.
Cited by
-
Reducing Peptide Sequence Bias in Quantitative Mass Spectrometry Data with Machine Learning.J Proteome Res. 2022 Jul 1;21(7):1771-1782. doi: 10.1021/acs.jproteome.2c00211. Epub 2022 Jun 13. J Proteome Res. 2022. PMID: 35696663 Free PMC article.
-
Evaluating nonpolar surface area and liquid chromatography/mass spectrometry response: an application for site occupancy measurements for enzyme intermediates in polyketide biosynthesis.Rapid Commun Mass Spectrom. 2014 Dec 15;28(23):2511-22. doi: 10.1002/rcm.7051. Rapid Commun Mass Spectrom. 2014. PMID: 25366398 Free PMC article.
-
Alkylating tryptic peptides to enhance electrospray ionization mass spectrometry analysis.Anal Chem. 2010 Dec 15;82(24):10135-42. doi: 10.1021/ac1019792. Epub 2010 Nov 29. Anal Chem. 2010. PMID: 21114270 Free PMC article.
-
Interplay of permanent charge and hydrophobicity in the electrospray ionization of glycans.Anal Chem. 2010 Aug 1;82(15):6636-42. doi: 10.1021/ac101227a. Anal Chem. 2010. PMID: 20590124 Free PMC article.
-
Hydrophobic derivatization of N-linked glycans for increased ion abundance in electrospray ionization mass spectrometry.J Am Soc Mass Spectrom. 2011 Aug;22(8):1309-17. doi: 10.1007/s13361-011-0140-x. Epub 2011 May 3. J Am Soc Mass Spectrom. 2011. PMID: 21953184 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

