Hydrophobic residues in helix 8 of cannabinoid receptor 1 are critical for structural and functional properties
- PMID: 20025243
- PMCID: PMC2904477
- DOI: 10.1021/bi901619r
Hydrophobic residues in helix 8 of cannabinoid receptor 1 are critical for structural and functional properties
Abstract
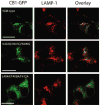
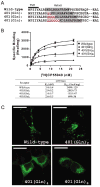
In addition to the heptahelical transmembrane domain shared by all G protein-coupled receptors (GPCRs), many class A GPCRs adopt a helical domain, termed helix 8, in the membrane-proximal region of the C terminus. We investigated the role of residues in the hydrophobic and hydrophilic faces of amphiphilic helix 8 of human cannabinoid receptor 1 (CB1). To differentiate between a role for specific residues and global features, we made two key mutants: one involving replacement of the highly hydrophobic groups, Leu404, Phe408, and Phe412, all with alanine and the second involving substitution of the basic residues, Lys402, Arg405, and Arg409, all with the neutral glutamine. The former showed a very low B(max) based on binding isotherms, a minimal E(max) based on GTPgammaS binding analysis, and defective localization relative to the wild-type CB1 receptor as revealed by confocal microscopy. However, the latter mutant and the wild-type receptors were indistinguishable. Circular dichroism spectroscopy of purified peptides with corresponding sequences indicated that the highly hydrophobic residues are critical for maintaining a strong helical structure in detergent, whereas the positively charged residues are not. Further investigation of mutant receptors revealed that CB1 localization requires a threshold level of hydrophobicity but not specific amino acids. Moreover, mutant receptors carrying two- to six-residue insertions amino-terminal to helix 8 revealed a graded decrease in B(max) values. Our results identify the key helix 8 components (including hydrophobicity of specific residues, structure, and location relative to TM7) determinant for receptor localization leading to robust ligand binding and G protein activation.
Figures
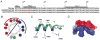
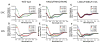


References
-
- Howlett AC, Barth F, Bonner TI, Cabral G, Casellas P, Devane WA, Felder CC, Herkenham M, Mackie K, Martin BR, Mechoulam R, Pertwee RG. International Union of Pharmacology. XXVII Classification of cannabinoid receptors. Pharmacol Rev. 2002;54:161–202. - PubMed
-
- Porter AC, Felder CC. The endocannabinoid nervous system: Unique opportunities for therapeutic intervention. Pharmacol Ther. 2001;90:45–60. - PubMed
-
- Gardner EL, Vorel SR. Cannabinoid transmission and reward-related events. Neurobiol Dis. 1998;5:502–533. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

