Mapping of the signal peptide-binding domain of Escherichia coli SecA using Förster resonance energy transfer
- PMID: 20025247
- PMCID: PMC2850574
- DOI: 10.1021/bi901446r
Mapping of the signal peptide-binding domain of Escherichia coli SecA using Förster resonance energy transfer
Abstract
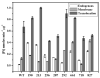
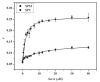
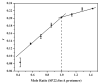
Identification of the signal peptide-binding domain within SecA ATPase is an important goal for understanding the molecular basis of SecA preprotein recognition as well as elucidating the chemo-mechanical cycle of this nanomotor during protein translocation. In this study, Forster resonance energy transfer methodology was employed to map the location of the SecA signal peptide-binding domain using a collection of functional monocysteine SecA mutants and alkaline phosphatase signal peptides labeled with appropriate donor-acceptor fluorophores. Fluorescence anisotropy measurements yielded an equilibrium binding constant of 1.4 or 10.7 muM for the alkaline phosphatase signal peptide labeled at residue 22 or 2, respectively, with SecA, and a binding stoichiometry of one signal peptide bound per SecA monomer. Binding affinity measurements performed with a monomer-biased mutant indicate that the signal peptide binds equally well to SecA monomer or dimer. Distance measurements determined for 13 SecA mutants show that the SecA signal peptide-binding domain encompasses a portion of the preprotein cross-linking domain but also includes regions of nucleotide-binding domain 1 and particularly the helical scaffold domain. The identified region lies at a multidomain interface within the heart of SecA, surrounded by and potentially responsive to domains important for binding nucleotide, mature portions of the preprotein, and the SecYEG channel. Our FRET-mapped binding domain, in contrast to the domain identified by NMR spectroscopy, includes the two-helix finger that has been shown to interact with the preprotein during translocation and lies at the entrance to the protein-conducting channel in the recently determined SecA-SecYEG structure.
Figures



Similar articles
-
Defining the solution state dimer structure of Escherichia coli SecA using Förster resonance energy transfer.Biochemistry. 2013 Apr 9;52(14):2388-401. doi: 10.1021/bi301217t. Epub 2013 Mar 29. Biochemistry. 2013. PMID: 23484952 Free PMC article.
-
Mapping of the SecA signal peptide binding site and dimeric interface by using the substituted cysteine accessibility method.J Bacteriol. 2013 Oct;195(20):4709-15. doi: 10.1128/JB.00661-13. Epub 2013 Aug 9. J Bacteriol. 2013. PMID: 23935053 Free PMC article.
-
Characterization of the Escherichia coli SecA signal peptide-binding site.J Bacteriol. 2012 Jan;194(2):307-16. doi: 10.1128/JB.06150-11. Epub 2011 Nov 4. J Bacteriol. 2012. PMID: 22056930 Free PMC article.
-
Oligomeric states of the SecA and SecYEG core components of the bacterial Sec translocon.Biochim Biophys Acta. 2007 Jan;1768(1):5-12. doi: 10.1016/j.bbamem.2006.08.013. Epub 2006 Aug 30. Biochim Biophys Acta. 2007. PMID: 17011510 Free PMC article. Review.
-
SecA-mediated targeting and translocation of secretory proteins.Biochim Biophys Acta. 2014 Aug;1843(8):1466-74. doi: 10.1016/j.bbamcr.2014.02.014. Epub 2014 Feb 25. Biochim Biophys Acta. 2014. PMID: 24583121 Review.
Cited by
-
Protein export by the mycobacterial SecA2 system is determined by the preprotein mature domain.J Bacteriol. 2013 Feb;195(4):672-81. doi: 10.1128/JB.02032-12. Epub 2012 Nov 30. J Bacteriol. 2013. PMID: 23204463 Free PMC article.
-
Fluorescence spectroscopy of soluble E. coli SPase I Δ2-75 reveals conformational changes in response to ligand binding.Proteins. 2014 Apr;82(4):596-606. doi: 10.1002/prot.24429. Epub 2013 Oct 17. Proteins. 2014. PMID: 24115229 Free PMC article.
-
A Specific interaction between SecA2 and a region of the preprotein adjacent to the signal peptide occurs during transport via the accessory Sec system.J Biol Chem. 2012 Jul 13;287(29):24438-47. doi: 10.1074/jbc.M112.378059. Epub 2012 May 31. J Biol Chem. 2012. PMID: 22654116 Free PMC article.
-
Conserved SecA Signal Peptide-Binding Site Revealed by Engineered Protein Chimeras and Förster Resonance Energy Transfer.Biochemistry. 2016 Mar 8;55(9):1291-300. doi: 10.1021/acs.biochem.5b01115. Epub 2016 Feb 19. Biochemistry. 2016. PMID: 26854513 Free PMC article.
-
Energetics of SecA dimerization.J Mol Biol. 2011 Apr 22;408(1):87-98. doi: 10.1016/j.jmb.2011.02.006. Epub 2011 Feb 15. J Mol Biol. 2011. PMID: 21315086 Free PMC article.
References
-
- Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 2008;77:643–667. - PubMed
-
- Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nat. Rev. Microbiol. 2007;5:839–851. - PubMed
-
- Papanikolau Y, Papadovasilaki M, Ravelli RB, McCarthy AA, Cusack S, Economou A, Petratos K. Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J. Mol. Biol. 2007;366:1545–1557. - PubMed
-
- Cabelli RJ, Dolan KM, Qian LP, Oliver DB. Characterization of membrane-associated and soluble states of SecA protein from wild-type and SecA51(TS) mutant strains of Escherichia coli. J. Biol. Chem. 1991;266:24420–24427. - PubMed
-
- Hartl FU, Lecker S, Schiebel E, Hendrick JP, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell. 1990;63:269–279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

