Expanding the family of collagen proteins: recombinant bacterial collagens of varying composition form triple-helices of similar stability
- PMID: 20025291
- PMCID: PMC2818787
- DOI: 10.1021/bm900894b
Expanding the family of collagen proteins: recombinant bacterial collagens of varying composition form triple-helices of similar stability
Abstract
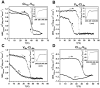
The presence of the (Gly-Xaa-Yaa)(n) open reading frames in different bacteria predicts the existence of an expanded family of collagen-like proteins. To further explore the triple-helix motif and stabilization mechanisms in the absence of hydroxyproline (Hyp), predicted novel collagen-like proteins from Gram-positive and -negative bacteria were expressed in Escherichia coli and characterized. Soluble proteins capable of successful folding and in vitro refolding were observed for collagen proteins from Methylobacterium sp 4-46, Rhodopseudomonas palustris and Solibacter usitatus . In contrast, all protein constructs from Clostridium perfringens were found predominantly in inclusion bodies. However, attachment of a heterologous N-terminal or C-terminal noncollagenous folding domain induced the Clostridium perfringens collagen domain to fold and become soluble. The soluble constructs from different bacteria had typical collagen triple-helical features and showed surprisingly similar thermal stabilities despite diverse amino acid compositions. These collagen-like proteins provide a resource for the development of biomaterials with new properties.
Figures




Similar articles
-
Bioengineered collagens: emerging directions for biomedical materials.Bioengineered. 2014 Jul-Aug;5(4):227-33. doi: 10.4161/bioe.28791. Epub 2014 Apr 9. Bioengineered. 2014. PMID: 24717980 Free PMC article. Review.
-
Noncollagenous region of the streptococcal collagen-like protein is a trimerization domain that supports refolding of adjacent homologous and heterologous collagenous domains.Protein Sci. 2010 Apr;19(4):775-85. doi: 10.1002/pro.356. Protein Sci. 2010. PMID: 20162611 Free PMC article.
-
Bacterial collagen-like proteins that form triple-helical structures.J Struct Biol. 2014 Jun;186(3):451-61. doi: 10.1016/j.jsb.2014.01.003. Epub 2014 Jan 14. J Struct Biol. 2014. PMID: 24434612 Free PMC article.
-
Mechanism of stabilization of a bacterial collagen triple helix in the absence of hydroxyproline.J Biol Chem. 2007 Oct 12;282(41):29757-65. doi: 10.1074/jbc.M703991200. Epub 2007 Aug 10. J Biol Chem. 2007. PMID: 17693404
-
The crucial role of trimerization domains in collagen folding.Int J Biochem Cell Biol. 2012 Jan;44(1):21-32. doi: 10.1016/j.biocel.2011.09.009. Epub 2011 Oct 5. Int J Biochem Cell Biol. 2012. PMID: 22001560 Review.
Cited by
-
Polymorphisms of a Collagen-Like Adhesin Contributes to Legionella pneumophila Adhesion, Biofilm Formation Capacity and Clinical Prevalence.Front Microbiol. 2019 Apr 5;10:604. doi: 10.3389/fmicb.2019.00604. eCollection 2019. Front Microbiol. 2019. PMID: 31024468 Free PMC article.
-
A Streptococcus pyogenes derived collagen-like protein as a non-cytotoxic and non-immunogenic cross-linkable biomaterial.Biomaterials. 2010 Apr;31(10):2755-61. doi: 10.1016/j.biomaterials.2009.12.040. Epub 2010 Jan 6. Biomaterials. 2010. PMID: 20056274 Free PMC article.
-
Bioengineered collagens: emerging directions for biomedical materials.Bioengineered. 2014 Jul-Aug;5(4):227-33. doi: 10.4161/bioe.28791. Epub 2014 Apr 9. Bioengineered. 2014. PMID: 24717980 Free PMC article. Review.
-
Designed coiled coils promote folding of a recombinant bacterial collagen.J Biol Chem. 2011 May 20;286(20):17512-20. doi: 10.1074/jbc.M110.217364. Epub 2011 Mar 28. J Biol Chem. 2011. PMID: 21454493 Free PMC article.
-
Recombinant Collagen Engineered to Bind to Discoidin Domain Receptor Functions as a Receptor Inhibitor.J Biol Chem. 2016 Feb 26;291(9):4343-55. doi: 10.1074/jbc.M115.674507. Epub 2015 Dec 23. J Biol Chem. 2016. PMID: 26702058 Free PMC article.
References
-
- Rasmussen M, Jacobsson M, Bjorck L. Genome-based identification and analysis of collagen-related structural motifs in bacterial and viral proteins. J Biol Chem. 2003;278(34):32313–6. - PubMed
-
- Ramachandran GN, Kartha G. Structure of collagen. Nature. 1955;176(4482):593–5. - PubMed
-
- Rich A, Crick FH. The molecular structure of collagen. J Mol Biol. 1961;3:483–506. - PubMed
-
- Jenkins CL, Raines RT. Insights on the conformational stability of collagen. Nat Prod Rep. 2002;19(1):49–59. - PubMed
-
- Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science. 1994;266(5182):75–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

