Overlapping repressor binding sites regulate expression of the Methanococcus maripaludis glnK(1) operon
- PMID: 20025661
- PMCID: PMC3104677
- DOI: 10.1111/j.1365-2958.2009.07016.x
Overlapping repressor binding sites regulate expression of the Methanococcus maripaludis glnK(1) operon
Abstract
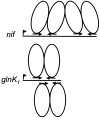
The euryarchaeal transcriptional repressor NrpR regulates a variety of nitrogen assimilation genes by 2-oxoglutarate-reversible binding to conserved palindromic operators. The number and positioning of these operators varies among promoter regions of regulated genes, suggesting NrpR can bind in different patterns. Particularly intriguing is the contrast between the nif and glnK(1) promoter regions of Methanococcus maripaludis, where two operators are present but with different configurations. Here we study NrpR binding and regulation at the glnK(1) promoter, where the two operator sequences overlap and occur on opposite faces of the double helix. We find that both operators function in binding, with a dimer of NrpR binding simultaneously to each overlapping operator. We show in vivo that the first operator plays a primary role in regulation and the second operator plays an enhancing role. This is the first demonstration of overlapping operators functioning in Archaea.
Figures



Similar articles
-
Genetic screen for regulatory mutations in Methanococcus maripaludis and its use in identification of induction-deficient mutants of the euryarchaeal repressor NrpR.Appl Environ Microbiol. 2007 Oct;73(20):6595-600. doi: 10.1128/AEM.01324-07. Epub 2007 Aug 24. Appl Environ Microbiol. 2007. PMID: 17720835 Free PMC article.
-
Regulation of nif expression in Methanococcus maripaludis: roles of the euryarchaeal repressor NrpR, 2-oxoglutarate, and two operators.J Biol Chem. 2005 Feb 18;280(7):5236-41. doi: 10.1074/jbc.M411778200. Epub 2004 Dec 7. J Biol Chem. 2005. PMID: 15590692
-
A novel repressor of nif and glnA expression in the methanogenic archaeon Methanococcus maripaludis.Mol Microbiol. 2003 Jan;47(1):235-46. doi: 10.1046/j.1365-2958.2003.03293.x. Mol Microbiol. 2003. PMID: 12492867
-
Metabolic processes of Methanococcus maripaludis and potential applications.Microb Cell Fact. 2016 Jun 10;15(1):107. doi: 10.1186/s12934-016-0500-0. Microb Cell Fact. 2016. PMID: 27286964 Free PMC article. Review.
-
Methanococcus maripaludis: an archaeon with multiple functional MCM proteins?Biochem Soc Trans. 2009 Feb;37(Pt 1):1-6. doi: 10.1042/BST0370001. Biochem Soc Trans. 2009. PMID: 19143592 Review.
Cited by
-
Contribution of transcriptomics to systems-level understanding of methanogenic Archaea.Archaea. 2013;2013:586369. doi: 10.1155/2013/586369. Epub 2013 Feb 27. Archaea. 2013. PMID: 23533330 Free PMC article. Review.
-
The complete genome sequence of the rumen methanogen Methanobacterium formicicum BRM9.Stand Genomic Sci. 2014 Dec 8;9:15. doi: 10.1186/1944-3277-9-15. eCollection 2014. Stand Genomic Sci. 2014. PMID: 25780506 Free PMC article.
-
Structural underpinnings of nitrogen regulation by the prototypical nitrogen-responsive transcriptional factor NrpR.Structure. 2010 Nov 10;18(11):1512-21. doi: 10.1016/j.str.2010.08.014. Structure. 2010. PMID: 21070950 Free PMC article.
-
Parallel evolution of transcriptome architecture during genome reorganization.Genome Res. 2011 Nov;21(11):1892-904. doi: 10.1101/gr.122218.111. Epub 2011 Jul 12. Genome Res. 2011. PMID: 21750103 Free PMC article.
-
Methanobacterium formicicum as a target rumen methanogen for the development of new methane mitigation interventions: A review.Vet Anim Sci. 2018 Sep 13;6:86-94. doi: 10.1016/j.vas.2018.09.001. eCollection 2018 Dec. Vet Anim Sci. 2018. PMID: 32734058 Free PMC article. Review.
References
-
- Augustus AM, Reardon PN, Heller WT, Spicer LD. Structural basis for the differential regulation of DNA by the methionine repressor MetJ. J Biol Chem. 2006;281:34269–34276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

