Direct MinE-membrane interaction contributes to the proper localization of MinDE in E. coli
- PMID: 20025670
- PMCID: PMC2814086
- DOI: 10.1111/j.1365-2958.2009.07006.x
Direct MinE-membrane interaction contributes to the proper localization of MinDE in E. coli
Abstract
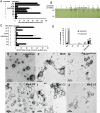
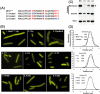
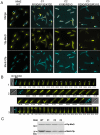
Dynamic oscillation of the Min system in Escherichia coli determines the placement of the division plane at the midcell. In addition to stimulating MinD ATPase activity, we report here that MinE can directly interact with the membrane and this interaction contributes to the proper MinDE localization and dynamics. The N-terminal domain of MinE is involved in direct contact between MinE and the membranes that may subsequently be stabilized by the C-terminal domain of MinE. In an in vitro system, MinE caused liposome deformation into membrane tubules, a property similar to that previously reported for MinD. We isolated a mutant MinE containing residue substitutions in R10, K11 and K12 that was fully capable of stimulating MinD ATPase activity, but was deficient in membrane binding. Importantly, this mutant was unable to support normal MinDE localization and oscillation, suggesting that direct MinE interaction with the membrane is critical for the dynamic behavior of the Min system.
Figures


References
-
- de Boer PAJ, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. - PubMed
-
- Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. - PubMed
-
- Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T. The proteomics of N-terminal methionine cleavage. Mol Cell Proteomics. 2006;5:2336–2349. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

