Isolation and characterization of a novel plasma membrane protein, osteoblast induction factor (obif), associated with osteoblast differentiation
- PMID: 20025746
- PMCID: PMC2805627
- DOI: 10.1186/1471-213X-9-70
Isolation and characterization of a novel plasma membrane protein, osteoblast induction factor (obif), associated with osteoblast differentiation
Abstract
Background: While several cell types are known to contribute to bone formation, the major player is a common bone matrix-secreting cell type, the osteoblast. Chondrocytes, which plays critical roles at several stages of endochondral ossification, and osteoblasts are derived from common precursors, and both intrinsic cues and signals from extrinsic cues play critical roles in the lineage decision of these cell types. Several studies have shown that cell fate commitment within the osteoblast lineage requires sequential, stage-specific signaling to promote osteoblastic differentiation programs. In osteoblastic differentiation, the functional mechanisms of transcriptional regulators have been well elucidated, however the exact roles of extrinsic molecules in osteoblastic differentiation are less clear.
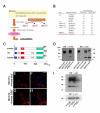
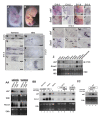
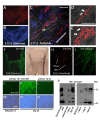
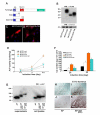
Results: We identify a novel gene, obif (osteoblast induction factor), encoding a transmembrane protein that is predominantly expressed in osteoblasts. During mouse development, obif is initially observed in the limb bud in a complementary pattern to Sox9 expression. Later in development, obif is highly expressed in osteoblasts at the stage of endochondral ossification. In cell line models, obif is up-regulated during osteoblastic differentiation. Exogenous obif expression stimulates osteoblastic differentiation and obif knockdown inhibits osteoblastic differentiation in preosteblastic MC3T3-E1 cells. In addition, the extracellular domain of obif protein exhibits functions similar to the full-length obif protein in induction of MC3T3-E1 differentiation.
Conclusions: Our results suggest that obif plays a role in osteoblastic differentiation by acting as a ligand.
Figures


Similar articles
-
OBIF, an osteoblast induction factor, plays an essential role in bone formation in association with osteoblastogenesis.Dev Growth Differ. 2012 May;54(4):474-80. doi: 10.1111/j.1440-169X.2012.01333.x. Epub 2012 Mar 15. Dev Growth Differ. 2012. PMID: 22416756
-
Obif, a Transmembrane Protein, Is Required for Bone Mineralization and Spermatogenesis in Mice.PLoS One. 2015 Jul 24;10(7):e0133704. doi: 10.1371/journal.pone.0133704. eCollection 2015. PLoS One. 2015. PMID: 26207632 Free PMC article.
-
Extracellular matrix-associated bone morphogenetic proteins are essential for differentiation of murine osteoblastic cells in vitro.Endocrinology. 1999 May;140(5):2125-33. doi: 10.1210/endo.140.5.6704. Endocrinology. 1999. PMID: 10218963
-
Akt activation is required for TGF-β1-induced osteoblast differentiation of MC3T3-E1 pre-osteoblasts.PLoS One. 2014 Dec 3;9(12):e112566. doi: 10.1371/journal.pone.0112566. eCollection 2014. PLoS One. 2014. PMID: 25470129 Free PMC article.
-
Ets transcription factors and targets in osteogenesis.Oncogene. 2000 Dec 18;19(55):6455-63. doi: 10.1038/sj.onc.1204037. Oncogene. 2000. PMID: 11175361 Review.
Cited by
-
FAD104, a regulatory factor of adipogenesis, acts as a novel regulator of calvarial bone formation.J Biol Chem. 2013 Nov 1;288(44):31772-83. doi: 10.1074/jbc.M113.452961. Epub 2013 Sep 19. J Biol Chem. 2013. PMID: 24052261 Free PMC article.
-
Parathyroid hormone-responsive Smad3-related factor, Tmem119, promotes osteoblast differentiation and interacts with the bone morphogenetic protein-Runx2 pathway.J Biol Chem. 2011 Mar 18;286(11):9787-96. doi: 10.1074/jbc.M110.179127. Epub 2011 Jan 14. J Biol Chem. 2011. PMID: 21239498 Free PMC article.
-
A systematic characterization of microglia-like cell occurrence during retinal organoid differentiation.iScience. 2022 Jun 11;25(7):104580. doi: 10.1016/j.isci.2022.104580. eCollection 2022 Jul 15. iScience. 2022. PMID: 35789843 Free PMC article.
-
The role of TMEM119 in gastric adenocarcinoma and its specific effects on immunity.J Int Med Res. 2025 Apr;53(4):3000605241306668. doi: 10.1177/03000605241306668. Epub 2025 Apr 12. J Int Med Res. 2025. PMID: 40219804 Free PMC article.
-
Microglia states and nomenclature: A field at its crossroads.Neuron. 2022 Nov 2;110(21):3458-3483. doi: 10.1016/j.neuron.2022.10.020. Neuron. 2022. PMID: 36327895 Free PMC article. Review.
References
-
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–71. doi: 10.1016/S0092-8674(00)80259-7. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

