Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu
- PMID: 20025795
- PMCID: PMC2832932
- DOI: 10.1017/S0033583509990060
Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu
Abstract
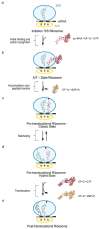
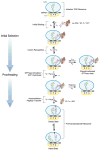
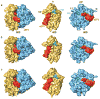
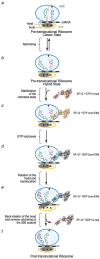
The ribosome is a complex macromolecular machine that translates the message encoded in the messenger RNA and synthesizes polypeptides by linking the individual amino acids carried by the cognate transfer RNAs (tRNAs). The protein elongation cycle, during which the tRNAs traverse the ribosome in a coordinated manner along a path of more than 100 A, is facilitated by large-scale rearrangements of the ribosome. These rearrangements go hand in hand with conformational changes of tRNA as well as elongation factors EF-Tu and EF-G - GTPases that catalyze tRNA delivery and translocation, respectively. This review focuses on the structural data related to the dynamics of the ribosomal machinery, which are the basis, in conjunction with existing biochemical, kinetic, and fluorescence resonance energy transfer data, of our knowledge of the decoding and translocation steps of protein elongation.
Figures
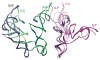





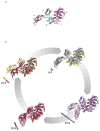
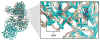

References
-
- Abel KM, Yoder MD, Hilgenfeld R, Jurnak F. An α to β conformational switch in EF-Tu. Structure. 1996;4:1153–1159. - PubMed
-
- Agrawal RK, Heagle AB, Penczek P, Grassucci R, Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nature Structural Biology. 1999;6:643–647. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

