New method for global alignment of 2 DNA sequences by the tree data structure
- PMID: 20025888
- PMCID: PMC7094160
- DOI: 10.1016/j.jtbi.2009.12.012
New method for global alignment of 2 DNA sequences by the tree data structure
Abstract
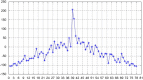
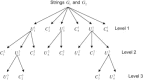
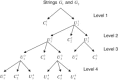
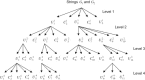
We introduce a new approach to investigate problem of DNA sequence alignment. The method consists of three parts: (i) simple alignment algorithm, (ii) extension algorithm for largest common substring, (iii) graphical simple alignment tree (GSA tree). The approach firstly obtains a graphical representation of scores of DNA sequences by the scoring equation R(0)*R-S(0)*S-T(0)*(a+bk). Then a GSA tree is constructed to facilitate solving the problem for global alignment of 2 DNA sequences. Finally we give several practical examples to illustrate the utility and practicality of the approach.
Crown Copyright 2009. Published by Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
FOGSAA: Fast Optimal Global Sequence Alignment Algorithm.Sci Rep. 2013;3:1746. doi: 10.1038/srep01746. Sci Rep. 2013. PMID: 23624407 Free PMC article.
-
DNA solution based on sequence alignment to the Minimum Spanning Tree problem.Int J Bioinform Res Appl. 2008;4(2):188-200. doi: 10.1504/IJBRA.2008.018345. Int J Bioinform Res Appl. 2008. PMID: 18490262
-
An Eulerian path approach to global multiple alignment for DNA sequences.J Comput Biol. 2003;10(6):803-19. doi: 10.1089/106652703322756096. J Comput Biol. 2003. PMID: 14980012
-
Numerical Characterization of DNA Sequences for Alignment-free Sequence Comparison - A Review.Comb Chem High Throughput Screen. 2022;25(3):365-380. doi: 10.2174/1386207324666210811101437. Comb Chem High Throughput Screen. 2022. PMID: 34382516 Review.
-
Finding homologs to nucleic acid or protein sequences using the framesearch program.Curr Protoc Bioinformatics. 2002 Aug;Chapter 3:Unit 3.2. doi: 10.1002/0471250953.bi0302s00. Curr Protoc Bioinformatics. 2002. PMID: 18792937 Review.
Cited by
-
Comparison of genomic data via statistical distribution.J Theor Biol. 2016 Oct 21;407:318-327. doi: 10.1016/j.jtbi.2016.07.032. Epub 2016 Jul 25. J Theor Biol. 2016. PMID: 27460589 Free PMC article.
-
Image correlation method for DNA sequence alignment.PLoS One. 2012;7(6):e39221. doi: 10.1371/journal.pone.0039221. Epub 2012 Jun 27. PLoS One. 2012. PMID: 22761742 Free PMC article.
References
-
- Althaus I.W., Chou J.J., Gonzales A.J., Diebel M.R., Chou K.C., Kezdy F.J., Romero D.L., Aristoff P.A., Tarpley W.G., Reusser F. Kinetic studies with the nonnucleoside HIV-1 reverse transcriptase inhibitor U-88204E. Biochemistry. 1993;32:6548–6554. - PubMed
-
- Althaus I.W., Chou J.J., Gonzales A.J., Diebel M.R., Chou K.C., Kezdy F.J., Romero D.L., Aristoff P.A., Tarpley W.G., Reusser F. Steady-state kinetic studies with the non-nucleoside HIV-1 reverse transcriptase inhibitor U-87201E. Journal of Biological Chemistry. 1993;268:6119–6124. - PubMed
-
- Althaus I.W., Chou J.J., Gonzales A.J., Diebel M.R., Chou K.C., Kezdy F.J., Romero D.L., Aristoff P.A., Tarpley W.G., Reusser F. The quinoline U-78036 is a potent inhibitor of HIV-1 reverse transcriptase. Journal of Biological Chemistry. 1993;268:14875–14880. - PubMed
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

