Role of the adenine ligand on the stabilization of the secondary and tertiary interactions in the adenine riboswitch
- PMID: 20026131
- PMCID: PMC2824916
- DOI: 10.1016/j.jmb.2009.12.024
Role of the adenine ligand on the stabilization of the secondary and tertiary interactions in the adenine riboswitch
Abstract
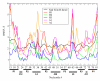
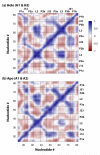
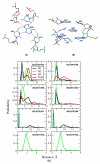
Riboswitches are RNA-based genetic control elements that function via a conformational transition mechanism when a specific target molecule binds to its binding pocket. To facilitate an atomic detail interpretation of experimental investigations on the role of the adenine ligand on the conformational properties and kinetics of folding of the add adenine riboswitch, we performed molecular dynamics simulations in both the presence and the absence of the ligand. In the absence of ligand, structural deviations were observed in the J23 junction and the P1 stem. Destabilization of the P1 stem in the absence of ligand involves the loss of direct stabilizing interactions with the ligand, with additional contributions from the J23 junction region. The J23 junction of the riboswitch is found to be more flexible, and the tertiary contacts among the junction regions are altered in the absence of the adenine ligand; results suggest that the adenine ligand associates and dissociates from the riboswitch in the vicinity of J23. Good agreement was obtained with the experimental data with the results indicating dynamic behavior of the adenine ligand on the nanosecond time scale to be associated with the dynamic behavior of hydrogen bonding with the riboswitch. Results also predict that direct interactions of the adenine ligand with U74 of the riboswitch are not essential for stable binding although it is crucial for its recognition. The possibility of methodological artifacts and force-field inaccuracies impacting the present observations was checked by additional molecular dynamics simulations in the presence of 2,6-diaminopurine and in the crystal environment.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
Molecular dynamics simulation of the binding process of ligands to the add adenine riboswitch aptamer.Phys Rev E. 2019 Aug;100(2-1):022412. doi: 10.1103/PhysRevE.100.022412. Phys Rev E. 2019. PMID: 31574664
-
Ligand-induced stabilization of the aptamer terminal helix in the add adenine riboswitch.RNA. 2013 Nov;19(11):1517-24. doi: 10.1261/rna.040493.113. Epub 2013 Sep 19. RNA. 2013. PMID: 24051105 Free PMC article.
-
Role of ligand binding in structural organization of add A-riboswitch aptamer: a molecular dynamics simulation.J Biomol Struct Dyn. 2011 Oct;29(2):403-16. doi: 10.1080/07391102.2011.10507394. J Biomol Struct Dyn. 2011. PMID: 21875158
-
[The adenine riboswitch: a new gene regulation mechanism].Med Sci (Paris). 2006 Dec;22(12):1053-9. doi: 10.1051/medsci/200622121053. Med Sci (Paris). 2006. PMID: 17156726 Review. French.
-
Riboswitch Mechanisms for Regulation of P1 Helix Stability.Int J Mol Sci. 2024 Oct 4;25(19):10682. doi: 10.3390/ijms251910682. Int J Mol Sci. 2024. PMID: 39409011 Free PMC article. Review.
Cited by
-
Binding free energy decomposition and multiple unbinding paths of buried ligands in a PreQ1 riboswitch.PLoS Comput Biol. 2021 Nov 12;17(11):e1009603. doi: 10.1371/journal.pcbi.1009603. eCollection 2021 Nov. PLoS Comput Biol. 2021. PMID: 34767553 Free PMC article.
-
Reduced model captures Mg(2+)-RNA interaction free energy of riboswitches.Biophys J. 2014 Apr 1;106(7):1508-19. doi: 10.1016/j.bpj.2014.01.042. Biophys J. 2014. PMID: 24703312 Free PMC article.
-
Elastic network models for RNA: a comparative assessment with molecular dynamics and SHAPE experiments.Nucleic Acids Res. 2015 Sep 3;43(15):7260-9. doi: 10.1093/nar/gkv708. Epub 2015 Jul 17. Nucleic Acids Res. 2015. PMID: 26187990 Free PMC article.
-
Recent Developments and Applications of the CHARMM force fields.Wiley Interdiscip Rev Comput Mol Sci. 2012 Jan;2(1):167-185. doi: 10.1002/wcms.74. Epub 2011 Jun 28. Wiley Interdiscip Rev Comput Mol Sci. 2012. PMID: 23066428 Free PMC article.
-
Kinetics of allosteric transitions in S-adenosylmethionine riboswitch are accurately predicted from the folding landscape.J Am Chem Soc. 2013 Nov 6;135(44):16641-50. doi: 10.1021/ja408595e. Epub 2013 Oct 22. J Am Chem Soc. 2013. PMID: 24087850 Free PMC article.
References
-
- Breaker RR. Complex riboswitches. Science. 2008;319:1795–1797. - PubMed
-
- Edwards TE, Klein DJ, Ferre-D’Amare AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Current Opinion in Structural Biology. 2007;17:273–279. - PubMed
-
- Kim JN, Breaker RR. Purine sensing by riboswitches. Biology of the Cell. 2008;100:1–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

