Genome-wide localization analysis of a complete set of Tafs reveals a specific effect of the taf1 mutation on Taf2 occupancy and provides indirect evidence for different TFIID conformations at different promoters
- PMID: 20026583
- PMCID: PMC2847235
- DOI: 10.1093/nar/gkp1172
Genome-wide localization analysis of a complete set of Tafs reveals a specific effect of the taf1 mutation on Taf2 occupancy and provides indirect evidence for different TFIID conformations at different promoters
Abstract
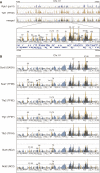
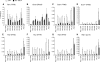
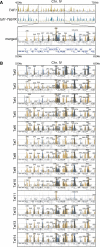
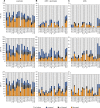
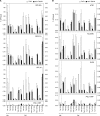
In Saccharomyces cerevisiae, TFIID and SAGA principally mediate transcription of constitutive housekeeping genes and stress-inducible genes, respectively, by delivering TBP to the core promoter. Both are multi-protein complexes composed of 15 and 20 subunits, respectively, five of which are common and which may constitute a core sub-module in each complex. Although genome-wide gene expression studies have been conducted extensively in several TFIID and/or SAGA mutants, there are only a limited number of studies investigating genome-wide localization of the components of these two complexes. Specifically, there are no previous reports on localization of a complete set of Tafs and the effects of taf mutations on localization. Here, we examine the localization profiles of a complete set of Tafs, Gcn5, Bur6/Ncb2, Sua7, Tfa2, Tfg1, Tfb3 and Rpb1, on chromosomes III, IV and V by chromatin immunoprecipitation (ChIP)-chip analysis in wild-type and taf1-T657K mutant strains. In addition, we conducted conventional and sequential ChIP analysis of several ribosomal protein genes (RPGs) and non-RPGs. Intriguingly, the results revealed a novel relationship between TFIIB and NC2, simultaneous co-localization of SAGA and TFIID on RPG promoters, specific effects of taf1 mutation on Taf2 occupancy, and an indirect evidence for the existence of different TFIID conformations.
Figures
Similar articles
-
The TAF9 C-terminal conserved region domain is required for SAGA and TFIID promoter occupancy to promote transcriptional activation.Mol Cell Biol. 2014 May;34(9):1547-63. doi: 10.1128/MCB.01060-13. Epub 2014 Feb 18. Mol Cell Biol. 2014. PMID: 24550006 Free PMC article.
-
Saccharomyces cerevisiae HMO1 interacts with TFIID and participates in start site selection by RNA polymerase II.Nucleic Acids Res. 2008 Mar;36(4):1343-57. doi: 10.1093/nar/gkm1068. Epub 2008 Jan 10. Nucleic Acids Res. 2008. PMID: 18187511 Free PMC article.
-
SAGA mediates transcription from the TATA-like element independently of Taf1p/TFIID but dependent on core promoter structures in Saccharomyces cerevisiae.PLoS One. 2017 Nov 27;12(11):e0188435. doi: 10.1371/journal.pone.0188435. eCollection 2017. PLoS One. 2017. PMID: 29176831 Free PMC article.
-
SAGA and TFIID: Friends of TBP drifting apart.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194604. doi: 10.1016/j.bbagrm.2020.194604. Epub 2020 Jul 14. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32673655 Review.
-
Architecture of the multi-functional SAGA complex and the molecular mechanism of holding TBP.FEBS J. 2021 May;288(10):3135-3147. doi: 10.1111/febs.15563. Epub 2020 Sep 29. FEBS J. 2021. PMID: 32946670 Review.
Cited by
-
Eaf1p Is Required for Recruitment of NuA4 in Targeting TFIID to the Promoters of the Ribosomal Protein Genes for Transcriptional Initiation In Vivo.Mol Cell Biol. 2015 Sep 1;35(17):2947-64. doi: 10.1128/MCB.01524-14. Epub 2015 Jun 22. Mol Cell Biol. 2015. PMID: 26100014 Free PMC article.
-
Nucleosome repositioning underlies dynamic gene expression.Genes Dev. 2016 Mar 15;30(6):660-72. doi: 10.1101/gad.274910.115. Epub 2016 Mar 10. Genes Dev. 2016. PMID: 26966245 Free PMC article.
-
A novel quantitative hemolytic assay coupled with restriction fragment length polymorphisms analysis enabled early diagnosis of atypical hemolytic uremic syndrome and identified unique predisposing mutations in Japan.PLoS One. 2015 May 7;10(5):e0124655. doi: 10.1371/journal.pone.0124655. eCollection 2015. PLoS One. 2015. PMID: 25951460 Free PMC article.
-
New insights into the function of transcription factor TFIID from recent structural studies.Curr Opin Genet Dev. 2011 Apr;21(2):219-24. doi: 10.1016/j.gde.2011.01.009. Epub 2011 Mar 21. Curr Opin Genet Dev. 2011. PMID: 21420851 Free PMC article. Review.
-
Sequential recruitment of SAGA and TFIID in a genomic response to DNA damage in Saccharomyces cerevisiae.Mol Cell Biol. 2011 Jan;31(1):190-202. doi: 10.1128/MCB.00317-10. Epub 2010 Oct 18. Mol Cell Biol. 2011. PMID: 20956559 Free PMC article.
References
-
- Mellor J. The dynamics of chromatin remodeling at promoters. Mol. Cell. 2005;19:147–157. - PubMed
-
- Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 2004;5:403–410. - PubMed
-
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. - PubMed
-
- Tora L. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 2002;16:673–675. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

