Effects of abasic sites on structural, thermodynamic and kinetic properties of quadruplex structures
- PMID: 20026588
- PMCID: PMC2847214
- DOI: 10.1093/nar/gkp1087
Effects of abasic sites on structural, thermodynamic and kinetic properties of quadruplex structures
Abstract
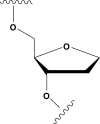
Abasic sites represent the most frequent lesion in DNA. Since several events generating abasic sites concern guanines, this damage is particularly important in quadruplex forming G-rich sequences, many of which are believed to be involved in several biological roles. However, the effects of abasic sites in sequences forming quadruplexes have been poorly studied. Here, we investigated the effects of abasic site mimics on structural, thermodynamic and kinetic properties of parallel quadruplexes. Investigation concerned five oligodeoxynucleotides based on the sequence d(TGGGGGT), in which all guanines have been replaced, one at a time, by an abasic site mimic (dS). All sequences preserve their ability to form quadruplexes; however, both spectroscopic and kinetic experiments point to sequence-dependent different effects on the structural flexibility and stability. Sequences d(TSGGGGT) and d(TGGGGST) form quite stable quadruplexes; however, for the other sequences, the introduction of the dS in proximity of the 3'-end decreases the stability more considerably than the 5'-end. Noteworthy, sequence d(TGSGGGT) forms a quadruplex where dS does not hamper the stacking between the G-tetrads adjacent to it. These results strongly argue for the central role of apurinic/apyrimidinic site damages and they encourage the production of further studies to better delineate the consequences of their presence in the biological relevant regions of the genome.
Figures


Similar articles
-
Riboflavin Stabilizes Abasic, Oxidized G-Quadruplex Structures.Biochemistry. 2022 Feb 15;61(4):265-275. doi: 10.1021/acs.biochem.1c00598. Epub 2022 Feb 1. Biochemistry. 2022. PMID: 35104101 Free PMC article.
-
The abasic site lesions in the human telomeric sequence d[TA(G(3)T(2)A)(3)G(3)]: a thermodynamic point of view.Biochim Biophys Acta. 2012 Dec;1820(12):2037-43. doi: 10.1016/j.bbagen.2012.09.011. Epub 2012 Sep 20. Biochim Biophys Acta. 2012. PMID: 23000492
-
Thermodynamics of dimerization of parallel quadruplex DNAs through the end-to-end stacking of 3'-terminal G-quartets.Nucleic Acids Symp Ser (Oxf). 2007;(51):245-6. doi: 10.1093/nass/nrm123. Nucleic Acids Symp Ser (Oxf). 2007. PMID: 18029678
-
Diverse effects of naturally occurring base lesions on the structure and stability of the human telomere DNA quadruplex.Biochimie. 2015 Nov;118:15-25. doi: 10.1016/j.biochi.2015.07.013. Epub 2015 Jul 15. Biochimie. 2015. PMID: 26188111 Review.
-
Assembly of chemically modified G-rich sequences into tetramolecular DNA G-quadruplexes and higher order structures.Methods. 2014 May 15;67(2):159-68. doi: 10.1016/j.ymeth.2014.01.004. Epub 2014 Jan 13. Methods. 2014. PMID: 24434505 Review.
Cited by
-
Endogenous oxidized DNA bases and APE1 regulate the formation of G-quadruplex structures in the genome.Proc Natl Acad Sci U S A. 2020 May 26;117(21):11409-11420. doi: 10.1073/pnas.1912355117. Epub 2020 May 13. Proc Natl Acad Sci U S A. 2020. PMID: 32404420 Free PMC article.
-
Human AP endonuclease inefficiently removes abasic sites within G4 structures compared to duplex DNA.Nucleic Acids Res. 2014 Jul;42(12):7708-19. doi: 10.1093/nar/gku417. Epub 2014 May 21. Nucleic Acids Res. 2014. PMID: 24848015 Free PMC article.
-
Human AP-endonuclease (Ape1) activity on telomeric G4 structures is modulated by acetylatable lysine residues in the N-terminal sequence.DNA Repair (Amst). 2019 Jan;73:129-143. doi: 10.1016/j.dnarep.2018.11.010. Epub 2018 Nov 22. DNA Repair (Amst). 2019. PMID: 30509560 Free PMC article.
-
The effects of molecular crowding on the structure and stability of g-quadruplexes with an abasic site.J Nucleic Acids. 2011;2011:857149. doi: 10.4061/2011/857149. Epub 2011 Sep 21. J Nucleic Acids. 2011. PMID: 21949901 Free PMC article.
-
Riboflavin Stabilizes Abasic, Oxidized G-Quadruplex Structures.Biochemistry. 2022 Feb 15;61(4):265-275. doi: 10.1021/acs.biochem.1c00598. Epub 2022 Feb 1. Biochemistry. 2022. PMID: 35104101 Free PMC article.
References
-
- Goljer I, Kumar S, Bolton PH. Refined solution structure of a DNA heteroduplex containing an aldehydic abasic site. J. Biol. Chem. 1995;270:22980–22987. - PubMed
-
- Loeb LA, Preston B. Mutagenesis by apurinic/apyrimidinic sites. Ann. Rev. Genet. 1996;20:201–230. - PubMed
-
- Lindahl T. DNA repair enzymes. Annu. Rev. Biochem. 1982;51:61–87. - PubMed
-
- Weiss B, Grossman L. Phosphodiesterases involved in DNA repair. Adv. Enzymol. Relat. Areas Mol. Biol. 1987;60:1–34. - PubMed
-
- McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: glycosylase mechanisms and structures. Annu. Rev. Biochem. 1999;68:255–285. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

