Structure and epigenetics of nucleoli in comparison with non-nucleolar compartments
- PMID: 20026667
- PMCID: PMC2857811
- DOI: 10.1369/jhc.2009.955435
Structure and epigenetics of nucleoli in comparison with non-nucleolar compartments
Abstract
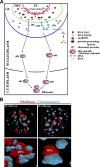
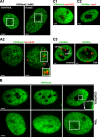
The nucleolus is a nuclear compartment that plays an important role in ribosome biogenesis. Some structural features and epigenetic patterns are shared between nucleolar and non-nucleolar compartments. For example, the location of transcriptionally active mRNA on extended chromatin loop species is similar to that observed for transcriptionally active ribosomal DNA (rDNA) genes on so-called Christmas tree branches. Similarly, nucleolus organizer region-bearing chromosomes located a distance from the nucleolus extend chromatin fibers into the nucleolar compartment. Specific epigenetic events, such as histone acetylation and methylation and DNA methylation, also regulate transcription of both rRNA- and mRNA-encoding loci. Here, we review the epigenetic mechanisms and structural features that regulate transcription of ribosomal and mRNA genes. We focus on similarities in epigenetic and structural regulation of chromatin in nucleoli and the surrounding non-nucleolar region and discuss the role of proteins, such as heterochromatin protein 1, fibrillarin, nucleolin, and upstream binding factor, in rRNA synthesis and processing.
Figures

Similar articles
-
Multiparameter microscopic analysis of nucleolar structure and ribosomal gene transcription.Histochem Cell Biol. 1996 Aug;106(2):167-92. doi: 10.1007/BF02484399. Histochem Cell Biol. 1996. PMID: 8877378 Review.
-
Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells.Nucleic Acids Res. 2007;35(7):2191-8. doi: 10.1093/nar/gkm118. Epub 2007 Mar 13. Nucleic Acids Res. 2007. PMID: 17355984 Free PMC article.
-
Loss of nucleolar histone chaperone NPM1 triggers rearrangement of heterochromatin and synergizes with a deficiency in DNA methyltransferase DNMT3A to drive ribosomal DNA transcription.J Biol Chem. 2014 Dec 12;289(50):34601-19. doi: 10.1074/jbc.M114.569244. Epub 2014 Oct 27. J Biol Chem. 2014. PMID: 25349213 Free PMC article.
-
Conditional inactivation of Upstream Binding Factor reveals its epigenetic functions and the existence of a somatic nucleolar precursor body.PLoS Genet. 2014 Aug 14;10(8):e1004505. doi: 10.1371/journal.pgen.1004505. eCollection 2014 Aug. PLoS Genet. 2014. PMID: 25121932 Free PMC article.
-
Integrating the genomic architecture of human nucleolar organizer regions with the biophysical properties of nucleoli.FEBS J. 2017 Dec;284(23):3977-3985. doi: 10.1111/febs.14108. Epub 2017 Jun 2. FEBS J. 2017. PMID: 28500793 Review.
Cited by
-
Nucleolar Reorganization Upon Site-Specific Double-Strand Break Induction.J Histochem Cytochem. 2016 Nov;64(11):669-686. doi: 10.1369/0022155416668505. Epub 2016 Sep 30. J Histochem Cytochem. 2016. PMID: 27680669 Free PMC article.
-
Inhibitor of Growth 4 (ING4) is a positive regulator of rRNA synthesis.Sci Rep. 2019 Nov 21;9(1):17235. doi: 10.1038/s41598-019-53767-1. Sci Rep. 2019. PMID: 31754246 Free PMC article.
-
Nucleolar sub-compartments in motion during rRNA synthesis inhibition: Contraction of nucleolar condensed chromatin and gathering of fibrillar centers are concomitant.PLoS One. 2017 Nov 30;12(11):e0187977. doi: 10.1371/journal.pone.0187977. eCollection 2017. PLoS One. 2017. PMID: 29190286 Free PMC article.
-
Altered inhibition in tuberous sclerosis and type IIb cortical dysplasia.Ann Neurol. 2012 Apr;71(4):539-51. doi: 10.1002/ana.22696. Epub 2012 Mar 23. Ann Neurol. 2012. PMID: 22447678 Free PMC article.
-
Stressing on the nucleolus in cardiovascular disease.Biochim Biophys Acta. 2014 Jun;1842(6):798-801. doi: 10.1016/j.bbadis.2013.09.016. Epub 2013 Oct 24. Biochim Biophys Acta. 2014. PMID: 24514103 Free PMC article. Review.
References
-
- Arabi A, Wu S, Ridderstråle K, Bierhoff H, Shiue C, Fatyol K, Fahlén S, et al. (2005) c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 7:303–310 - PubMed
-
- Bártová E, Harničarová A, Krejčí J, Strašák L, Kozubek S (2008a) Single-cell c-myc gene expression in relationship to nuclear domains. Chromosome Res 16:325–343 - PubMed
-
- Bártová E, Kozubek S (2006) Nuclear architecture in the light of gene expression and cell differentiation studies. Biol Cell 98:323–336 - PubMed
-
- Bártová E, Krejčí J, Harničarová A, Kozubek S (2008c) Differentiation of human embryonic stem cells induces condensation of chromosome territories and formation of heterochromatin protein 1 foci. Differentiation 76:24–32 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

