Regulation by blue light of the fluffy gene encoding a major regulator of conidiation in Neurospora crassa
- PMID: 20026679
- PMCID: PMC2845335
- DOI: 10.1534/genetics.109.109975
Regulation by blue light of the fluffy gene encoding a major regulator of conidiation in Neurospora crassa
Abstract
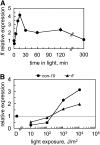
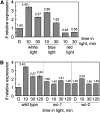
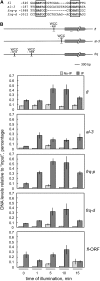
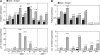
The development of asexual spores, that is, the process of conidiation, in the fungus Neurospora crassa is increased by light. The fluffy (fl) gene, encoding a major regulator of conidiation, is activated by light. We describe here a detailed characterization of the regulation by blue light of fl in vegetative hyphae. This induction requires the white collar complex (WCC) while the FLD protein acts as a dark repressor of fl transcription. We show that the WCC directly regulates fl transcription in response to blue light after transiently binding the promoter. We propose that fl is repressed by FLD in vegetative mycelia and that the repression is lost after light exposure and WCC activation. The increase in fl mRNA in vegetative mycelia after light exposure, and the corresponding increase in the amount of the regulatory FL protein, should promote the activation of the conidiation pathway. The activation by light of fl provides a simple mechanism for the activation of conidiation by blue light in Neurospora that may be at work in other fungi.
Figures
Similar articles
-
Transcriptional Regulation by the Velvet Protein VE-1 during Asexual Development in the Fungus Neurospora crassa.mBio. 2022 Aug 30;13(4):e0150522. doi: 10.1128/mbio.01505-22. Epub 2022 Aug 1. mBio. 2022. PMID: 35913159 Free PMC article.
-
A complex photoreceptor system mediates the regulation by light of the conidiation genes con-10 and con-6 in Neurospora crassa.Fungal Genet Biol. 2010 Apr;47(4):352-63. doi: 10.1016/j.fgb.2009.11.004. Epub 2009 Nov 20. Fungal Genet Biol. 2010. PMID: 19932184
-
Conidiation in Neurospora crassa: vegetative reproduction by a model fungus.Int Microbiol. 2020 Jan;23(1):97-105. doi: 10.1007/s10123-019-00085-1. Epub 2019 Jun 6. Int Microbiol. 2020. PMID: 31172300 Review.
-
Fluffy, the major regulator of conidiation in Neurospora crassa, directly activates a developmentally regulated hydrophobin gene.Mol Microbiol. 2005 Apr;56(1):282-97. doi: 10.1111/j.1365-2958.2005.04544.x. Mol Microbiol. 2005. PMID: 15773996
-
Light input and processing in the circadian clock of Neurospora.FEBS Lett. 2011 May 20;585(10):1467-73. doi: 10.1016/j.febslet.2011.03.050. Epub 2011 Mar 29. FEBS Lett. 2011. PMID: 21453703 Review.
Cited by
-
The social network: deciphering fungal language.Nat Rev Microbiol. 2011 Jun;9(6):440-51. doi: 10.1038/nrmicro2580. Nat Rev Microbiol. 2011. PMID: 21572459 Review.
-
Construction of Light-Responsive Gene Regulatory Network for Growth, Development and Secondary Metabolite Production in Cordyceps militaris.Biology (Basel). 2022 Jan 4;11(1):71. doi: 10.3390/biology11010071. Biology (Basel). 2022. PMID: 35053069 Free PMC article.
-
Alteration of light-dependent gene regulation by the absence of the RCO-1/RCM-1 repressor complex in the fungus Neurospora crassa.PLoS One. 2014 Apr 18;9(4):e95069. doi: 10.1371/journal.pone.0095069. eCollection 2014. PLoS One. 2014. PMID: 24747913 Free PMC article.
-
Identification of the transcription factor Znc1p, which regulates the yeast-to-hypha transition in the dimorphic yeast Yarrowia lipolytica.PLoS One. 2013 Jun 24;8(6):e66790. doi: 10.1371/journal.pone.0066790. Print 2013. PLoS One. 2013. PMID: 23826133 Free PMC article.
-
Genome-wide characterization of light-regulated genes in Neurospora crassa.G3 (Bethesda). 2014 Jul 21;4(9):1731-45. doi: 10.1534/g3.114.012617. G3 (Bethesda). 2014. PMID: 25053707 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

