A small, variable, and irregular killer cell Ig-like receptor locus accompanies the absence of MHC-C and MHC-G in gibbons
- PMID: 20026738
- PMCID: PMC3017126
- DOI: 10.4049/jimmunol.0903016
A small, variable, and irregular killer cell Ig-like receptor locus accompanies the absence of MHC-C and MHC-G in gibbons
Abstract
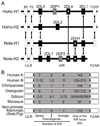
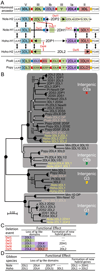
The killer cell Ig-like receptors (KIRs) of NK cells recognize MHC class I ligands and function in placental reproduction and immune defense against pathogens. During the evolution of monkeys, great apes, and humans, an ancestral KIR3DL gene expanded to become a diverse and rapidly evolving gene family of four KIR lineages. Characterizing the KIR locus are three framework regions, defining two intervals of variable gene content. By analysis of four KIR haplotypes from two species of gibbon, we find that the smaller apes do not conform to these rules. Although diverse and irregular in structure, the gibbon haplotypes are unusually small, containing only two to five functional genes. Comparison with the predicted ancestral hominoid KIR haplotype indicates that modern gibbon KIR haplotypes were formed by a series of deletion events, which created new hybrid genes as well as eliminating ancestral genes. Of the three framework regions, only KIR3DL3 (lineage V), defining the 5' end of the KIR locus, is present and intact on all gibbon KIR haplotypes. KIR2DL4 (lineage I) defining the central framework region has been a major target for elimination or inactivation, correlating with the absence of its putative ligand, MHC-G, in gibbons. Similarly, the MHC-C-driven expansion of lineage III KIR genes in great apes has not occurred in gibbons because they lack MHC-C. Our results indicate that the selective forces shaping the size and organization of the gibbon KIR locus differed from those acting upon the KIR of other hominoid species.
Figures




Similar articles
-
Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules.Philos Trans R Soc Lond B Biol Sci. 2012 Mar 19;367(1590):800-11. doi: 10.1098/rstb.2011.0266. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22312047 Free PMC article. Review.
-
Evolution of killer cell Ig-like receptor (KIR) genes: definition of an orangutan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC-C.J Immunol. 2007 Jul 1;179(1):491-504. doi: 10.4049/jimmunol.179.1.491. J Immunol. 2007. PMID: 17579070
-
Definition of the cattle killer cell Ig-like receptor gene family: comparison with aurochs and human counterparts.J Immunol. 2014 Dec 15;193(12):6016-30. doi: 10.4049/jimmunol.1401980. Epub 2014 Nov 14. J Immunol. 2014. PMID: 25398326 Free PMC article.
-
Co-evolution of MHC class I and variable NK cell receptors in placental mammals.Immunol Rev. 2015 Sep;267(1):259-82. doi: 10.1111/imr.12326. Immunol Rev. 2015. PMID: 26284483 Free PMC article. Review.
-
Evolutionary patterns of killer cell Ig-like receptor genes in Old World monkeys.Gene. 2011 Mar 15;474(1-2):39-51. doi: 10.1016/j.gene.2010.12.006. Epub 2010 Dec 24. Gene. 2011. PMID: 21185924
Cited by
-
Resurrecting KIR2DP1: A Key Intermediate in the Evolution of Human Inhibitory NK Cell Receptors That Recognize HLA-C.J Immunol. 2017 Mar 1;198(5):1961-1973. doi: 10.4049/jimmunol.1601835. Epub 2017 Jan 25. J Immunol. 2017. PMID: 28122963 Free PMC article.
-
KIR2DL5: An Orphan Inhibitory Receptor Displaying Complex Patterns of Polymorphism and Expression.Front Immunol. 2012 Sep 17;3:289. doi: 10.3389/fimmu.2012.00289. eCollection 2012. Front Immunol. 2012. PMID: 23060877 Free PMC article.
-
Two Orangutan Species Have Evolved Different KIR Alleles and Haplotypes.J Immunol. 2017 Apr 15;198(8):3157-3169. doi: 10.4049/jimmunol.1602163. Epub 2017 Mar 6. J Immunol. 2017. PMID: 28264973 Free PMC article.
-
Variable NK cell receptors exemplified by human KIR3DL1/S1.J Immunol. 2011 Jul 1;187(1):11-9. doi: 10.4049/jimmunol.0902332. J Immunol. 2011. PMID: 21690332 Free PMC article. Review.
-
Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules.Philos Trans R Soc Lond B Biol Sci. 2012 Mar 19;367(1590):800-11. doi: 10.1098/rstb.2011.0266. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22312047 Free PMC article. Review.
References
-
- Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu. Rev. Immunol. 2002;20:217–251. - PubMed
-
- Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu. Rev. Genomics Hum. Genet. 2006;7:277–300. - PubMed
-
- Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME, O'Brien SJ, Rosenberg WM, Thomas DL, Carrington M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. - PubMed
-
- Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, Lifson JD, Allen TM, Carrington M, Altfeld M. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J. Exp. Med. 2007;204:3027–3036. - PMC - PubMed
-
- Martin M, Qi Y, Gao X, Yamada E, Martin J, Pereyra F, Colombo S, Brown E, Shupert W, Phair J, Goedert J, Buchbinder S, Kirk G, Telenti A, Connors M, O'brien S, Walker B, Parham P, Deeks S, McVicar D, Carrington M. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007;39:733–740. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

