Expression of the VP2 protein of murine norovirus by a translation termination-reinitiation strategy
- PMID: 20027307
- PMCID: PMC2793014
- DOI: 10.1371/journal.pone.0008390
Expression of the VP2 protein of murine norovirus by a translation termination-reinitiation strategy
Abstract
Background: Expression of the minor virion structural protein VP2 of the calicivirus murine norovirus (MNV) is believed to occur by the unusual mechanism of termination codon-dependent reinitiation of translation. In this process, following translation of an upstream open reading frame (ORF) and termination at the stop codon, a proportion of 40S subunits remain associated with the mRNA and reinitiate at the AUG of a downstream ORF, which is typically in close proximity. Consistent with this, the VP2 start codon (AUG) of MNV overlaps the stop codon of the upstream VP1 ORF (UAA) in the pentanucleotide UAAUG.
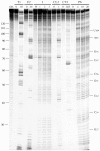
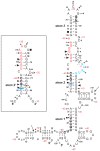
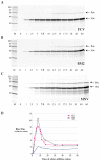
Principal findings: Here, we confirm that MNV VP2 expression is regulated by termination-reinitiation and define the mRNA sequence requirements. Efficient reintiation is dependent upon 43 nt of RNA immediately upstream of the UAAUG site. Chemical and enzymatic probing revealed that the RNA in this region is not highly structured and includes an essential stretch of bases complementary to 18S rRNA helix 26 (Motif 1). The relative position of Motif 1 with respect to the UAAUG site impacts upon the efficiency of the process. Termination-reinitiation in MNV was also found to be relatively insensitive to the initiation inhibitor edeine.
Conclusions: The termination-reinitiation signal of MNV most closely resembles that of influenza BM2. Similar to other viruses that use this strategy, base-pairing between mRNA and rRNA is likely to play a role in tethering the 40S subunit to the mRNA following termination at the VP1 stop codon. Our data also indicate that accurate recognition of the VP2 ORF AUG is not a pre-requisite for efficient reinitiation of translation in this system.
Conflict of interest statement
Figures





Similar articles
-
Characterization of the termination-reinitiation strategy employed in the expression of influenza B virus BM2 protein.RNA. 2008 Nov;14(11):2394-406. doi: 10.1261/rna.1231008. Epub 2008 Sep 29. RNA. 2008. PMID: 18824510 Free PMC article.
-
Further characterisation of the translational termination-reinitiation signal of the influenza B virus segment 7 RNA.PLoS One. 2011 Feb 8;6(2):e16822. doi: 10.1371/journal.pone.0016822. PLoS One. 2011. PMID: 21347434 Free PMC article.
-
Translational termination-reinitiation in RNA viruses.Biochem Soc Trans. 2010 Dec;38(6):1558-64. doi: 10.1042/BST0381558. Biochem Soc Trans. 2010. PMID: 21118126 Review.
-
The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation.Genes Dev. 2007 Dec 1;21(23):3149-62. doi: 10.1101/gad.439507. Genes Dev. 2007. PMID: 18056426 Free PMC article.
-
Does eIF3 promote reinitiation after translation of short upstream ORFs also in mammalian cells?RNA Biol. 2017 Dec 2;14(12):1660-1667. doi: 10.1080/15476286.2017.1353863. Epub 2017 Sep 15. RNA Biol. 2017. PMID: 28745933 Free PMC article. Review.
Cited by
-
Major Capsid Protein Synthesis from the Genomic RNA of Feline Calicivirus.J Virol. 2020 Jul 16;94(15):e00280-20. doi: 10.1128/JVI.00280-20. Print 2020 Jul 16. J Virol. 2020. PMID: 32404528 Free PMC article.
-
Two alternative ways of start site selection in human norovirus reinitiation of translation.J Biol Chem. 2014 Apr 25;289(17):11739-11754. doi: 10.1074/jbc.M114.554030. Epub 2014 Mar 5. J Biol Chem. 2014. PMID: 24599949 Free PMC article.
-
A conserved class of viral RNA structures regulates translation reinitiation through dynamic ribosome interactions.Cell Rep. 2025 Feb 25;44(2):115236. doi: 10.1016/j.celrep.2025.115236. Epub 2025 Feb 1. Cell Rep. 2025. PMID: 39893634 Free PMC article.
-
Norovirus RNA synthesis is modulated by an interaction between the viral RNA-dependent RNA polymerase and the major capsid protein, VP1.J Virol. 2012 Sep;86(18):10138-49. doi: 10.1128/JVI.01208-12. Epub 2012 Jul 11. J Virol. 2012. Retraction in: J Virol. 2017 Nov 30;91(24):e01708-17. doi: 10.1128/JVI.01708-17. PMID: 22787222 Free PMC article. Retracted.
-
Structure-function relationship in the 'termination upstream ribosomal binding site' of the calicivirus rabbit hemorrhagic disease virus.Nucleic Acids Res. 2019 Feb 28;47(4):1920-1934. doi: 10.1093/nar/gkz021. Nucleic Acids Res. 2019. PMID: 30668745 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

