Residues essential for plasminogen binding by the cation-independent mannose 6-phosphate receptor
- PMID: 20028034
- PMCID: PMC2814163
- DOI: 10.1021/bi901779p
Residues essential for plasminogen binding by the cation-independent mannose 6-phosphate receptor
Abstract
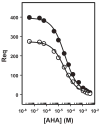
The 300 kDa cation-independent mannose 6-phosphate receptor (CI-MPR) is a multifunctional protein that binds diverse intracellular and extracellular ligands with high affinity. The CI-MPR is a receptor for plasminogen, and this interaction can be inhibited by lysine analogues. To characterize the molecular basis for this interaction, surface plasmon resonance (SPR) analyses were performed using truncated forms of the CI-MPR and plasminogen. The results show that the N-terminal region of the CI-MPR containing domains 1 and 2, but not domain 1 alone, of the receptor's 15-domain extracytoplasmic region binds plasminogen (K(d) = 5 +/- 1 nM) with an affinity similar to that of the full-length receptor (K(d) = 20 +/- 6 nM). In addition to its C-terminal serine protease domain, plasminogen contains lysine binding sites (LBS), which are located within each of its five kringle domains, except kringle 3. We show that kringles 1-4, but not kringles 1-3, bind the CI-MPR, indicating an essential role for the LBS in kringle 4 of plasminogen. To identify the lysine residue(s) of the CI-MPR that serve(s) as an essential determinant for recognition by the LBS of plasminogen, site-directed mutagenesis studies were carried out using a construct encoding the N-terminal three domains of the CI-MPR (Dom1-3His) which contains both a mannose 6-phosphate (Man-6-P) and plasminogen binding site. The results demonstrate two lysine residues (Lys53 located in domain 1 and Lys125 located in the loop connecting domains 1 and 2) of the CI-MPR are key determinants for plasminogen binding but are not required for Man-6-P binding.
Figures







Similar articles
-
Identification of a fourth mannose 6-phosphate binding site in the cation-independent mannose 6-phosphate receptor.Glycobiology. 2015 Jun;25(6):591-606. doi: 10.1093/glycob/cwv001. Epub 2015 Jan 8. Glycobiology. 2015. PMID: 25573276 Free PMC article.
-
Identification of a low affinity mannose 6-phosphate-binding site in domain 5 of the cation-independent mannose 6-phosphate receptor.J Biol Chem. 2004 Sep 10;279(37):38658-67. doi: 10.1074/jbc.M407474200. Epub 2004 Jul 12. J Biol Chem. 2004. PMID: 15252023
-
Domain 5 of the cation-independent mannose 6-phosphate receptor preferentially binds phosphodiesters (mannose 6-phosphate N-acetylglucosamine ester).Biochemistry. 2007 Nov 6;46(44):12604-17. doi: 10.1021/bi7011806. Epub 2007 Oct 10. Biochemistry. 2007. PMID: 17927214
-
The kringle domains of human plasminogen.Ciba Found Symp. 1997;212:46-60; discussion 60-5. doi: 10.1002/9780470515457.ch4. Ciba Found Symp. 1997. PMID: 9524763 Review.
-
Strategies for carbohydrate recognition by the mannose 6-phosphate receptors.Glycobiology. 2008 Sep;18(9):664-78. doi: 10.1093/glycob/cwn061. Epub 2008 Jul 11. Glycobiology. 2008. PMID: 18621992 Free PMC article. Review.
Cited by
-
Dissecting mannose 6-phosphate-insulin-like growth factor 2 receptor complexes that control activation and uptake of plasminogen in cells.J Biol Chem. 2012 Jun 29;287(27):22450-62. doi: 10.1074/jbc.M112.339663. Epub 2012 May 21. J Biol Chem. 2012. PMID: 22613725 Free PMC article.
-
Lysosomal enzyme binding to the cation-independent mannose 6-phosphate receptor is regulated allosterically by insulin-like growth factor 2.Sci Rep. 2024 Nov 6;14(1):26875. doi: 10.1038/s41598-024-75300-9. Sci Rep. 2024. PMID: 39505925 Free PMC article.
-
Expression of insulin-like growth factor 2 receptor in corneal keratocytes during differentiation and in response to wound healing.Invest Ophthalmol Vis Sci. 2014 Oct 30;55(12):7697-708. doi: 10.1167/iovs.14-15179. Invest Ophthalmol Vis Sci. 2014. PMID: 25358730 Free PMC article.
-
Mannose 6-phosphate receptor homology (MRH) domain-containing lectins in the secretory pathway.Biochim Biophys Acta. 2011 Sep;1810(9):815-26. doi: 10.1016/j.bbagen.2011.06.016. Epub 2011 Jun 24. Biochim Biophys Acta. 2011. PMID: 21723917 Free PMC article. Review.
-
Identification of a fourth mannose 6-phosphate binding site in the cation-independent mannose 6-phosphate receptor.Glycobiology. 2015 Jun;25(6):591-606. doi: 10.1093/glycob/cwv001. Epub 2015 Jan 8. Glycobiology. 2015. PMID: 25573276 Free PMC article.
References
-
- Tordai H, Banyai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999;461:63–67. - PubMed
-
- Castellino FJ, Ploplis VA. Structure and function of the plasminogen/plasmin system. Thromb Haemost. 2005;93:647–654. - PubMed
-
- Sehl LC, Castellino FJ. Thermodynamic properties of the binding of alpha-, omega-amino acids to the isolated kringle 4 region of human plasminogen as determined by high sensitivity titration calorimetry. J Biol Chem. 1990;265:5482–5486. - PubMed
-
- Hoover GJ, Menhart N, Martin A, Warder S, Castellino FJ. Amino acids of the recombinant kringle 1 domain of human plasminogen that stabilize its interaction with omega-amino acids. Biochemistry. 1993;32:10936–10943. - PubMed
-
- Menhart N, Castellino FJ. The importance of the hydrophobic components of the binding energies in the interaction of omega-amino acid ligands with isolated kringle polypeptide domains of human plasminogen. Int J Pept Protein Res. 1995;46:464–470. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

