Born-Oppenheimer ab initio QM/MM molecular dynamics simulations of the hydrolysis reaction catalyzed by protein arginine deiminase 4
- PMID: 20028143
- PMCID: PMC2801900
- DOI: 10.1021/jp9080614
Born-Oppenheimer ab initio QM/MM molecular dynamics simulations of the hydrolysis reaction catalyzed by protein arginine deiminase 4
Abstract
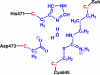
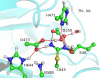
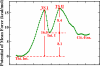
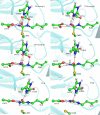
Protein arginine deiminase 4 (PAD4) catalyzes the citrullination of the peptidylarginine via two successive stages: deimination and hydrolysis. Herein, by employing state-of-the-art Born-Oppenheimer ab initio QM/MM molecular dynamics simulations with the umbrella sampling method, we characterized the catalytic mechanism of the hydrolysis reaction: first, the nucleophilic attack of a water molecule to the C(zeta) of the thiouronium intermediate yields a stable tetrahedral intermediate, and then the S-C(zeta) bond breaks to generate the final product, citrulline. Throughout the hydrolysis reaction, His471 and Asp473 play pivotal catalytic roles by first enhancing the nucleophilic ability of the active water through forming shorter and low-barrier hydrogen bonds and then by serving as proton-accepting groups to deprotonate the water molecule, which is consistent with experimental findings. At the transition state, the spontaneous proton transfer among the reactive water, His471 and Asp473 have been observed. The determined overall free energy barrier for this hydrolysis stage is 16.5 kcal x mol(-1), which is lower than the barrier of 20.9 kcal x mol(-1) for the deimination stage determined previously with the same computational approach [J. Phys. Chem. B 2009, 113, 12750-12758]. Thus, the rate-determining step of the PAD4-catalyzed citrullination is the first step of the deimination. Our current work further demonstrates the strength and applicability of the ab initio QM/MM MD approach in simulating enzyme reactions.
Figures

Similar articles
-
Active site cysteine is protonated in the PAD4 Michaelis complex: evidence from Born-Oppenheimer ab initio QM/MM molecular dynamics simulations.J Phys Chem B. 2009 Sep 24;113(38):12750-8. doi: 10.1021/jp903173c. J Phys Chem B. 2009. PMID: 19507815 Free PMC article.
-
Theoretical insights into the protonation states of active site cysteine and citrullination mechanism of Porphyromonas gingivalis peptidylarginine deiminase.Proteins. 2017 Aug;85(8):1518-1528. doi: 10.1002/prot.25313. Epub 2017 May 25. Proteins. 2017. PMID: 28486790
-
Catalytic reaction mechanism of acetylcholinesterase determined by Born-Oppenheimer ab initio QM/MM molecular dynamics simulations.J Phys Chem B. 2010 Jul 8;114(26):8817-25. doi: 10.1021/jp104258d. J Phys Chem B. 2010. PMID: 20550161 Free PMC article.
-
Peptidylarginine deiminase 4 and citrullination in health and disease.Autoimmun Rev. 2010 Jan;9(3):158-60. doi: 10.1016/j.autrev.2009.06.002. Epub 2009 Jun 18. Autoimmun Rev. 2010. PMID: 19540364 Review.
-
Peptidylarginine deiminases in citrullination, gene regulation, health and pathogenesis.Biochim Biophys Acta. 2013 Oct;1829(10):1126-35. doi: 10.1016/j.bbagrm.2013.07.003. Epub 2013 Jul 13. Biochim Biophys Acta. 2013. PMID: 23860259 Free PMC article. Review.
Cited by
-
Mechanistic studies of agmatine deiminase from multiple bacterial species.Biochemistry. 2010 Nov 2;49(43):9413-23. doi: 10.1021/bi101405y. Biochemistry. 2010. PMID: 20939536 Free PMC article.
-
Insight into the phosphodiesterase mechanism from combined QM/MM free energy simulations.FEBS J. 2011 Jul;278(14):2579-95. doi: 10.1111/j.1742-4658.2011.08187.x. Epub 2011 Jun 14. FEBS J. 2011. PMID: 21595828 Free PMC article.
-
Why does the G117H mutation considerably improve the activity of human butyrylcholinesterase against sarin? Insights from quantum mechanical/molecular mechanical free energy calculations.Biochemistry. 2012 Nov 6;51(44):8980-92. doi: 10.1021/bi3009246. Epub 2012 Oct 23. Biochemistry. 2012. PMID: 23092211 Free PMC article.
-
How is acetylcholinesterase phosphonylated by soman? An ab initio QM/MM molecular dynamics study.J Phys Chem A. 2014 Oct 2;118(39):9132-9. doi: 10.1021/jp502712d. Epub 2014 May 9. J Phys Chem A. 2014. PMID: 24786171 Free PMC article.
-
Interrogation of the Active Sites of Protein Arginine Deiminases (PAD1, -2, and -4) Using Designer Probes.ACS Med Chem Lett. 2013 Jan 15;4(2):249-53. doi: 10.1021/ml300377d. eCollection 2013 Feb 14. ACS Med Chem Lett. 2013. PMID: 24900657 Free PMC article.
References
-
- Hagiwara T, Nakashima K, Hirano H, Senshu T, Yamada M. Biochem. Biophys. Res. Commun. 2002;290:979–983. - PubMed
-
- Nakashima K, Hagiwara T, Yamada M. J. Biol. Chem. 2002;277:49562–49568. - PubMed
-
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Science. 2004;306:279–283. - PubMed
-
- Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, Kouzarides T. Cell. 2004;118:545–553. - PubMed
-
- Hagiwara T, Hidaka Y, Yamada M. Biochemistry. 2005;44:5827–5834. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

