Expanding the definition of the classical bipartite nuclear localization signal
- PMID: 20028483
- PMCID: PMC2886731
- DOI: 10.1111/j.1600-0854.2009.01028.x
Expanding the definition of the classical bipartite nuclear localization signal
Abstract
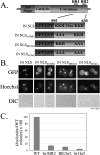
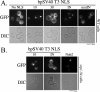
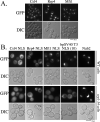
Nuclear localization signals (NLSs) are amino acid sequences that target cargo proteins into the nucleus. Rigorous characterization of NLS motifs is essential to understanding and predicting pathways for nuclear import. The best-characterized NLS is the classical NLS (cNLS), which is recognized by the cNLS receptor, importin-alpha. cNLSs are conventionally defined as having one (monopartite) or two clusters of basic amino acids separated by a 9-12 aa linker (bipartite). Motivated by the finding that Ty1 integrase, which contains an unconventional putative bipartite cNLS with a 29 aa linker, exploits the classical nuclear import machinery, we assessed the functional boundaries for linker length within a bipartite cNLS. We confirmed that the integrase cNLS is a bona fide bipartite cNLS, then carried out a systematic analysis of linker length in an obligate bipartite cNLS cargo, which revealed that some linkers longer than conventionally defined can function in nuclear import. Linker function is dependent on the sequence and likely the inherent flexibility of the linker. Subsequently, we interrogated the Saccharomyces cerevisiae proteome to identify cellular proteins containing putative long bipartite cNLSs. We experimentally confirmed that Rrp4 contains a bipartite cNLS with a 25 aa linker. Our studies show that the traditional definition of bipartite cNLSs is too restrictive and linker length can vary depending on amino acid composition.
Figures

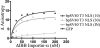

Similar articles
-
Classical NLS proteins from Saccharomyces cerevisiae.J Mol Biol. 2008 Jun 13;379(4):678-94. doi: 10.1016/j.jmb.2008.04.038. Epub 2008 Apr 22. J Mol Biol. 2008. PMID: 18485366
-
A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin α-dependent nuclear localization signals.Protein Sci. 2024 Feb;33(2):e4876. doi: 10.1002/pro.4876. Protein Sci. 2024. PMID: 38108201 Free PMC article.
-
A PY-NLS nuclear targeting signal is required for nuclear localization and function of the Saccharomyces cerevisiae mRNA-binding protein Hrp1.J Biol Chem. 2008 May 9;283(19):12926-34. doi: 10.1074/jbc.M800898200. Epub 2008 Mar 14. J Biol Chem. 2008. PMID: 18343812 Free PMC article.
-
Bipartite nuclear localization sequence is indispensable for nuclear import and stability of self-dimerization of ADARa in Bombyx mori.Insect Biochem Mol Biol. 2024 Nov;174:104190. doi: 10.1016/j.ibmb.2024.104190. Epub 2024 Oct 9. Insect Biochem Mol Biol. 2024. PMID: 39389319 Review.
-
Classical nuclear localization signals: definition, function, and interaction with importin alpha.J Biol Chem. 2007 Feb 23;282(8):5101-5. doi: 10.1074/jbc.R600026200. Epub 2006 Dec 14. J Biol Chem. 2007. PMID: 17170104 Free PMC article. Review.
Cited by
-
Recent advances in drug delivery systems for targeting brain tumors.Drug Deliv. 2023 Dec;30(1):1-18. doi: 10.1080/10717544.2022.2154409. Drug Deliv. 2023. PMID: 36597214 Free PMC article. Review.
-
Functional analysis of nuclear localization signals in VP1-2 homologues from all herpesvirus subfamilies.J Virol. 2014 May;88(10):5391-405. doi: 10.1128/JVI.03797-13. Epub 2014 Feb 26. J Virol. 2014. PMID: 24574406 Free PMC article.
-
Unconventional Pathways of Secretion Contribute to Inflammation.Int J Mol Sci. 2017 Jan 5;18(1):102. doi: 10.3390/ijms18010102. Int J Mol Sci. 2017. PMID: 28067797 Free PMC article. Review.
-
Nuclear Localization of the DNA Repair Scaffold XRCC1: Uncovering the Functional Role of a Bipartite NLS.Sci Rep. 2015 Aug 25;5:13405. doi: 10.1038/srep13405. Sci Rep. 2015. PMID: 26304019 Free PMC article.
-
Dual activity inhibition of threonine aspartase 1 by a single bisphosphate ligand.RSC Adv. 2022 Nov 30;12(53):34176-34184. doi: 10.1039/d2ra06019a. eCollection 2022 Nov 29. RSC Adv. 2022. PMID: 36545626 Free PMC article.
References
-
- Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci. 2000;113(2000):1651–1659. - PubMed
-
- Sorokin A, Kim E, Ovchinnikov L. Nucleocytoplasmic transport of proteins. Biochemistry (Mosc) 2007;72(13):1439–1457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

