Progressive colonization and restricted gene flow shape island-dependent population structure in Galápagos marine iguanas (Amblyrhynchus cristatus)
- PMID: 20028547
- PMCID: PMC2807874
- DOI: 10.1186/1471-2148-9-297
Progressive colonization and restricted gene flow shape island-dependent population structure in Galápagos marine iguanas (Amblyrhynchus cristatus)
Abstract
Background: Marine iguanas (Amblyrhynchus cristatus) inhabit the coastlines of large and small islands throughout the Galápagos archipelago, providing a rich system to study the spatial and temporal factors influencing the phylogeographic distribution and population structure of a species. Here, we analyze the microevolution of marine iguanas using the complete mitochondrial control region (CR) as well as 13 microsatellite loci representing more than 1200 individuals from 13 islands.
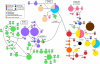
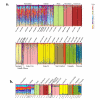
Results: CR data show that marine iguanas occupy three general clades: one that is widely distributed across the northern archipelago, and likely spread from east to west by way of the South Equatorial current, a second that is found mostly on the older eastern and central islands, and a third that is limited to the younger northern and western islands. Generally, the CR haplotype distribution pattern supports the colonization of the archipelago from the older, eastern islands to the younger, western islands. However, there are also signatures of recurrent, historical gene flow between islands after population establishment. Bayesian cluster analysis of microsatellite genotypes indicates the existence of twenty distinct genetic clusters generally following a one-cluster-per-island pattern. However, two well-differentiated clusters were found on the easternmost island of San Cristóbal, while nine distinct and highly intermixed clusters were found on youngest, westernmost islands of Isabela and Fernandina. High mtDNA and microsatellite genetic diversity were observed for populations on Isabela and Fernandina that may be the result of a recent population expansion and founder events from multiple sources.
Conclusions: While a past genetic study based on pure FST analysis suggested that marine iguana populations display high levels of nuclear (but not mitochondrial) gene flow due to male-biased dispersal, the results of our sex-biased dispersal tests and the finding of strong genetic differentiation between islands do not support this view. Therefore, our study is a nice example of how recently developed analytical tools such as Bayesian clustering analysis and DNA sequence-based demographic analyses can overcome potential biases introduced by simply relying on FST estimates from markers with different inheritance patterns.
Figures

Similar articles
-
Genetic differentiation between marine iguanas from different breeding sites on the island of Santa Fe (Galapagos Archipelago).J Hered. 2010 Nov-Dec;101(6):663-75. doi: 10.1093/jhered/esq067. Epub 2010 Jun 10. J Hered. 2010. PMID: 20538757
-
The complete mitochondrial genomes of the Galápagos iguanas, Amblyrhynchus cristatus and Conolophus subcristatus.Mitochondrial DNA A DNA Mapp Seq Anal. 2016 Sep;27(5):3699-700. doi: 10.3109/19401736.2015.1079863. Epub 2015 Sep 10. Mitochondrial DNA A DNA Mapp Seq Anal. 2016. PMID: 26357924
-
Genomic insights into the biogeography and evolution of Galápagos iguanas.Mol Phylogenet Evol. 2025 Mar;204:108294. doi: 10.1016/j.ympev.2025.108294. Epub 2025 Jan 27. Mol Phylogenet Evol. 2025. PMID: 39880223
-
Genetic variation in the invasive avian parasite, Philornis downsi (Diptera, Muscidae) on the Galápagos archipelago.BMC Ecol. 2008 Jul 31;8:13. doi: 10.1186/1472-6785-8-13. BMC Ecol. 2008. PMID: 18671861 Free PMC article.
-
Evolution of body size in Galapagos marine iguanas.Proc Biol Sci. 2005 Oct 7;272(1576):1985-93. doi: 10.1098/rspb.2005.3205. Proc Biol Sci. 2005. PMID: 16191607 Free PMC article. Review.
Cited by
-
Ecological and evolutionary influences on body size and shape in the Galápagos marine iguana (Amblyrhynchus cristatus).Oecologia. 2016 Jul;181(3):885-94. doi: 10.1007/s00442-016-3618-1. Epub 2016 Apr 4. Oecologia. 2016. PMID: 27041683
-
Salmonella strains isolated from Galápagos iguanas show spatial structuring of serovar and genomic diversity.PLoS One. 2012;7(5):e37302. doi: 10.1371/journal.pone.0037302. Epub 2012 May 16. PLoS One. 2012. PMID: 22615968 Free PMC article.
-
What Darwin could not see: island formation and historical sea levels shape genetic divergence and island biogeography in a coastal marine species.Heredity (Edinb). 2023 Sep;131(3):189-200. doi: 10.1038/s41437-023-00635-4. Epub 2023 Jul 3. Heredity (Edinb). 2023. PMID: 37400518 Free PMC article.
-
Genetic structure at three spatial scales is consistent with limited philopatry in Ricord's Rock Iguanas (Cyclura ricordii).Ecol Evol. 2019 Jul 2;9(14):8331-8350. doi: 10.1002/ece3.5414. eCollection 2019 Jul. Ecol Evol. 2019. PMID: 31380093 Free PMC article.
-
Hybridization masks speciation in the evolutionary history of the Galápagos marine iguana.Proc Biol Sci. 2015 Jun 22;282(1809):20150425. doi: 10.1098/rspb.2015.0425. Proc Biol Sci. 2015. PMID: 26041359 Free PMC article.
References
-
- Grant PR, Grant BR. How and why species multiply: the radiation of Darwin's finches. Princeton: Princeton University Press; 2008.
-
- Cox A. In: Patterns of Galápagos organisms. Bowman RI, Berson M, Leviton AE, editor. San Francisco: Pacific Division, AAAS; 1983. Ages of the Galápagos Islands; pp. 11–23.
-
- Arbogast BS, Drovetski SV, Curry RL, Boag PT, Seutin G, Grant PR, Grant BR, Anderson DJ. The origin and diversification of Galápagos mockingbirds. Evolution. 2006;60:370–382. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

