Potential impact of stress activated retrotransposons on genome evolution in a marine diatom
- PMID: 20028555
- PMCID: PMC2806351
- DOI: 10.1186/1471-2164-10-624
Potential impact of stress activated retrotransposons on genome evolution in a marine diatom
Abstract
Background: Transposable elements (TEs) are mobile DNA sequences present in the genomes of most organisms. They have been extensively studied in animals, fungi, and plants, and have been shown to have important functions in genome dynamics and species evolution. Recent genomic data can now enlarge the identification and study of TEs to other branches of the eukaryotic tree of life. Diatoms, which belong to the heterokont group, are unicellular eukaryotic algae responsible for around 40% of marine primary productivity. The genomes of a centric diatom, Thalassiosira pseudonana, and a pennate diatom, Phaeodactylum tricornutum, that likely diverged around 90 Mya, have recently become available.
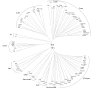
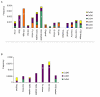
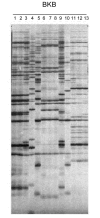
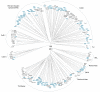
Results: In the present work, we establish that LTR retrotransposons (LTR-RTs) are the most abundant TEs inhabiting these genomes, with a much higher presence in the P. tricornutum genome. We show that the LTR-RTs found in diatoms form two new phylogenetic lineages that appear to be diatom specific and are also found in environmental samples taken from different oceans. Comparative expression analysis in P. tricornutum cells cultured under 16 different conditions demonstrate high levels of transcriptional activity of LTR retrotransposons in response to nitrate limitation and upon exposure to diatom-derived reactive aldehydes, which are known to induce stress responses and cell death. Regulatory aspects of P. tricornutum retrotransposon transcription also include the occurrence of nitrate limitation sensitive cis-regulatory components within LTR elements and cytosine methylation dynamics. Differential insertion patterns in different P. tricornutum accessions isolated from around the world infer the role of LTR-RTs in generating intraspecific genetic variability.
Conclusion: Based on these findings we propose that LTR-RTs may have been important for promoting genome rearrangements in diatoms.
Figures






Similar articles
-
The Phaeodactylum genome reveals the evolutionary history of diatom genomes.Nature. 2008 Nov 13;456(7219):239-44. doi: 10.1038/nature07410. Epub 2008 Oct 15. Nature. 2008. PMID: 18923393
-
Re-examination of two diatom reference genomes using long-read sequencing.BMC Genomics. 2021 May 24;22(1):379. doi: 10.1186/s12864-021-07666-3. BMC Genomics. 2021. PMID: 34030633 Free PMC article.
-
Complex repeat structures and novel features in the mitochondrial genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana.Gene. 2011 May 1;476(1-2):20-6. doi: 10.1016/j.gene.2011.02.001. Epub 2011 Feb 12. Gene. 2011. PMID: 21320580
-
LTR-retrotransposons in plants: Engines of evolution.Gene. 2017 Aug 30;626:14-25. doi: 10.1016/j.gene.2017.04.051. Epub 2017 May 2. Gene. 2017. PMID: 28476688 Review.
-
Co-evolution of plant LTR-retrotransposons and their host genomes.Protein Cell. 2013 Jul;4(7):493-501. doi: 10.1007/s13238-013-3037-6. Epub 2013 Jun 23. Protein Cell. 2013. PMID: 23794032 Free PMC article. Review.
Cited by
-
Protocol: Chromatin immunoprecipitation (ChIP) methodology to investigate histone modifications in two model diatom species.Plant Methods. 2012 Dec 7;8(1):48. doi: 10.1186/1746-4811-8-48. Plant Methods. 2012. PMID: 23217141 Free PMC article.
-
Conserved structure and inferred evolutionary history of long terminal repeats (LTRs).Mob DNA. 2013 Feb 1;4(1):5. doi: 10.1186/1759-8753-4-5. Mob DNA. 2013. PMID: 23369192 Free PMC article.
-
Strain-specific transcriptional responses overshadow salinity effects in a marine diatom sampled along the Baltic Sea salinity cline.ISME J. 2022 Jul;16(7):1776-1787. doi: 10.1038/s41396-022-01230-x. Epub 2022 Apr 5. ISME J. 2022. PMID: 35383290 Free PMC article.
-
Characterization of the small RNA transcriptome of the diatom, Thalassiosira pseudonana.PLoS One. 2011;6(8):e22870. doi: 10.1371/journal.pone.0022870. Epub 2011 Aug 12. PLoS One. 2011. PMID: 21857960 Free PMC article.
-
The model diatom Phaeodactylum tricornutum provides insights into the diversity and function of microeukaryotic DNA methyltransferases.Commun Biol. 2023 Mar 9;6(1):253. doi: 10.1038/s42003-023-04629-0. Commun Biol. 2023. PMID: 36894681 Free PMC article.
References
-
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. - PubMed
-
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, Ashburner M, Celniker SE. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3:RESEARCH0084. doi: 10.1186/gb-2002-3-12-research0084. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

