NAD(P)H quinone-oxydoreductase 1 protects eukaryotic translation initiation factor 4GI from degradation by the proteasome
- PMID: 20028737
- PMCID: PMC2815573
- DOI: 10.1128/MCB.00868-09
NAD(P)H quinone-oxydoreductase 1 protects eukaryotic translation initiation factor 4GI from degradation by the proteasome
Abstract
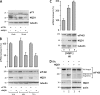
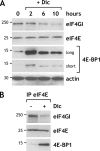
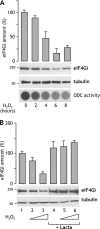
The eukaryotic translation initiation factor 4GI (eIF4GI) serves as a central adapter in cap-binding complex assembly. Although eIF4GI has been shown to be sensitive to proteasomal degradation, how the eIF4GI steady-state level is controlled remains unknown. Here, we show that eIF4GI exists in a complex with NAD(P)H quinone-oxydoreductase 1 (NQO1) in cell extracts. Treatment of cells with dicumarol (dicoumarol), a pharmacological inhibitor of NQO1 known to preclude NQO1 binding to its protein partners, provokes eIF4GI degradation by the proteasome. Consistently, the eIF4GI steady-state level also diminishes upon the silencing of NQO1 (by transfection with small interfering RNA), while eIF4GI accumulates upon the overexpression of NQO1 (by transfection with cDNA). We further reveal that treatment of cells with dicumarol frees eIF4GI from mRNA translation initiation complexes due to strong activation of its natural competitor, the translational repressor 4E-BP1. As a consequence of cap-binding complex dissociation and eIF4GI degradation, protein synthesis is dramatically inhibited. Finally, we show that the regulation of eIF4GI stability by the proteasome may be prominent under oxidative stress. Our findings assign NQO1 an original role in the regulation of mRNA translation via the control of eIF4GI stability by the proteasome.
Figures

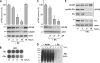


References
-
- Asher, G., Z. Bercovich, P. Tsvetkov, Y. Shaul, and C. Kahana. 2005. 20S proteasomal degradation of ornithine decarboxylase is regulated by NQO1. Mol. Cell 17:645-655. - PubMed
-
- Baugh, J. M., and E. V. Pilipenko. 2004. 20S proteasome differentially alters translation of different mRNAs via the cleavage of eIF4F and eIF3. Mol. Cell 16:575-586. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
