Identification of clusterin domain involved in NF-kappaB pathway regulation
- PMID: 20028970
- PMCID: PMC2836031
- DOI: 10.1074/jbc.C109.057133
Identification of clusterin domain involved in NF-kappaB pathway regulation
Abstract
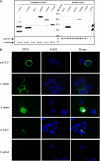
Clusterin (CLU) is a ubiquitous protein that has been implicated in tumorigenesis, apoptosis, inflammation, and cell proliferation. We and others have previously shown that CLU is an inhibitor of the NF-kappaB pathway. However, the exact form of CLU and the region(s) of CLU involved in this effect were unknown. Using newly generated molecular constructs encoding for CLU and various regions of the molecule, we demonstrated that the presecretory form of CLU (psCLU) form bears the NF-kappaB regulatory activity. Sequence comparison analysis showed sequence motif identity between CLU and beta-transducin repeat-containing protein (beta-TrCP), a main E3 ubiquitin ligase involved in IkappaB-alpha degradation. These homologies were localized in the disulfide constraint region of CLU. We generated a specific molecular construct of this region, named DeltaCLU, and showed that it has the same NF-kappaB regulatory activity as CLU. Neither the alpha-chain nor the beta-chain of CLU had any NF-kappaB regulatory activity. Furthermore, we showed that following tumor necrosis factor-alpha stimulation of transfected cells, we could co-immunoprecipitate phospho-IkappaB-alpha with DeltaCLU. Moreover, we showed that DeltaCLU could localize both in the cytoplasm and in the nucleus. These results demonstrate the identification of a new CLU activity site involved in NF-kappaB pathway regulation.
Figures

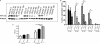
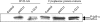
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

