Targets of the Tal1 transcription factor in erythrocytes: E2 ubiquitin conjugase regulation by Tal1
- PMID: 20028976
- PMCID: PMC2820762
- DOI: 10.1074/jbc.M109.030296
Targets of the Tal1 transcription factor in erythrocytes: E2 ubiquitin conjugase regulation by Tal1
Abstract
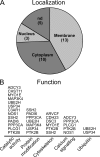
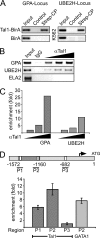
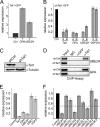
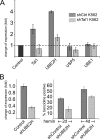
The Tal1 transcription factor is essential for the development of the hematopoietic system and plays a role during definitive erythropoiesis in the adult. Despite the importance of Tal1 in erythropoiesis, only a small number of erythroid differentiation target genes are known. A chromatin precipitation and cloning approach was established to uncover novel Tal1 target genes in erythropoiesis. The BirA tag/BirA ligase biotinylation system in combination with streptavidin chromatin precipitation (Strep-CP) was used to co-precipitate genomic DNA bound to Tal1. Tal1 was found to bind in the vicinity of 31 genes including the E2-ubiquitin conjugase UBE2H gene. Binding of Tal1 to UBE2H was confirmed by chromatin immunoprecipitation. UBE2H expression is increased during erythroid differentiation of hCD34(+) cells. Tal1 expression activated UBE2H expression, whereas Tal1 knock-down reduced UBE2H expression and ubiquitin transfer activity. This study identifies parts of the ubiquitinylation machinery as a cellular target downstream of the transcription factor Tal1 and provides novel insights into Tal1-regulated erythropoiesis.
Figures


Similar articles
-
ID3 promotes erythroid differentiation and is repressed by a TAL1-PRMT6 complex.J Biol Chem. 2025 Feb;301(2):108119. doi: 10.1016/j.jbc.2024.108119. Epub 2024 Dec 22. J Biol Chem. 2025. PMID: 39716491 Free PMC article.
-
Characterization of DNA-binding-dependent and -independent functions of SCL/TAL1 during human erythropoiesis.Blood. 2004 May 1;103(9):3326-35. doi: 10.1182/blood-2003-05-1689. Epub 2004 Jan 8. Blood. 2004. PMID: 14715640
-
Genome-wide identification of TAL1's functional targets: insights into its mechanisms of action in primary erythroid cells.Genome Res. 2010 Aug;20(8):1064-83. doi: 10.1101/gr.104935.110. Epub 2010 Jun 21. Genome Res. 2010. PMID: 20566737 Free PMC article.
-
Transcription Factor TAL1 in Erythropoiesis.Adv Exp Med Biol. 2024;1459:243-258. doi: 10.1007/978-3-031-62731-6_11. Adv Exp Med Biol. 2024. PMID: 39017847 Review.
-
Stem Cell Leukemia: how a TALented actor can go awry on the hematopoietic stage.Leukemia. 2016 Oct;30(10):1968-1978. doi: 10.1038/leu.2016.169. Epub 2016 Jun 13. Leukemia. 2016. PMID: 27443261 Review.
Cited by
-
Oncogenic transcriptional program driven by TAL1 in T-cell acute lymphoblastic leukemia.Int J Hematol. 2019 Jan;109(1):5-17. doi: 10.1007/s12185-018-2518-z. Epub 2018 Aug 25. Int J Hematol. 2019. PMID: 30145780 Review.
-
Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart.Proc Natl Acad Sci U S A. 2011 Apr 5;108(14):5632-7. doi: 10.1073/pnas.1016959108. Epub 2011 Mar 17. Proc Natl Acad Sci U S A. 2011. PMID: 21415370 Free PMC article.
-
Modular UBE2H-CTLH E2-E3 complexes regulate erythroid maturation.Elife. 2022 Dec 2;11:e77937. doi: 10.7554/eLife.77937. Elife. 2022. PMID: 36459484 Free PMC article.
-
Bayesian information sharing enhances detection of regulatory associations in rare cell types.Bioinformatics. 2021 Jul 12;37(Suppl_1):i349-i357. doi: 10.1093/bioinformatics/btab269. Bioinformatics. 2021. PMID: 34252956 Free PMC article.
-
Exome sequencing identifies novel genetic variants associated with varicose veins.PLoS Genet. 2024 Jul 9;20(7):e1011339. doi: 10.1371/journal.pgen.1011339. eCollection 2024 Jul. PLoS Genet. 2024. PMID: 38980841 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

