Interaction between Hhex and SOX13 modulates Wnt/TCF activity
- PMID: 20028982
- PMCID: PMC2820800
- DOI: 10.1074/jbc.M109.046649
Interaction between Hhex and SOX13 modulates Wnt/TCF activity
Abstract
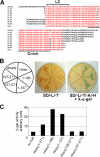
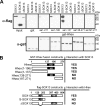
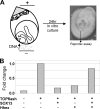
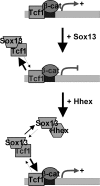
Fine-tuning of the Wnt/TCF pathway is crucial for multiple embryological processes, including liver development. Here we describe how the interaction between Hhex (hematopoietically expressed homeobox) and SOX13 (SRY-related high mobility group box transcription factor 13), modulates Wnt/TCF pathway activity. Hhex is a homeodomain factor expressed in multiple endoderm-derived tissues, like the liver, where it is essential for proper development. The pleiotropic expression of Hhex during embryonic development and its dual role as a transcriptional repressor and activator suggest the presence of different tissue-specific partners capable of modulating its activity and function. While searching for developmentally regulated Hhex partners, we set up a yeast two-hybrid screening using an E9.5-10.5 mouse embryo library and the N-terminal domain of Hhex as bait. Among the putative protein interactors, we selected SOX13 for further characterization. We found that SOX13 interacts directly with full-length Hhex, and we delineated the interaction domains within the two proteins. SOX13 is known to repress Wnt/TCF signaling by interacting with TCF1. We show that Hhex is able to block the SOX13-dependent repression of Wnt/TCF activity by displacing SOX13 from the SOX13 x TCF1 complex. Moreover, Hhex de-repressed the Wnt/TCF pathway in the ventral foregut endoderm of cultured mouse embryos electroporated with a SOX13-expressing plasmid. We conclude that the interaction between Hhex and SOX13 may contribute to control Wnt/TCF signaling in the early embryo.
Figures






References
-
- McLin V. A., Rankin S. A., Zorn A. M. (2007) Development 134, 2207–2217 - PubMed
-
- Ober E. A., Verkade H., Field H. A., Stainier D. Y. (2006) Nature 442, 688–691 - PubMed
-
- Thomas P. Q., Brown A., Beddington R. S. (1998) Development 125, 85–94 - PubMed
-
- Bort R., Martinez-Barbera J. P., Beddington R. S., Zaret K. S. (2004) Development 131, 797–806 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

