Cbl controls EGFR fate by regulating early endosome fusion
- PMID: 20029031
- PMCID: PMC2813709
- DOI: 10.1126/scisignal.2000217
Cbl controls EGFR fate by regulating early endosome fusion
Abstract
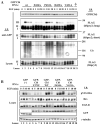
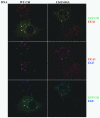
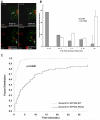
Amino acid residues 1 to 434 of the E3 ubiquitin ligase Cbl control signaling of the epidermal growth factor receptor (EGFR) by enhancing its ubiquitination, down-regulation, and lysosomal degradation. This region of Cbl comprises a tyrosine kinase-binding domain, a linker region, a really interesting new gene finger (RF), and a subset of the residues of the RF tail. In experiments with full-length alanine substitution mutants, we demonstrated that the RF tail of Cbl regulated biochemically distinct checkpoints in the endocytosis of EGFR. The Cbl- and ubiquitin-dependent degradation of the regulator of internalization hSprouty2 was compromised by the Val(431)--> Ala mutation, whereas the Cbl- and EGFR-dependent dephosphorylation or degradation of the endosomal trafficking regulator Hrs was compromised by the Phe(434)--> Ala mutation. Deregulated phosphorylation of Hrs correlated with inhibition of the fusion of early endosomes and of the degradation of EGFR. This study provides the first evidence that Cbl regulates receptor fate by controlling the fusion of sorting endosomes. We postulate that it does so by modulating the abundance of tyrosine-phosphorylated Hrs.
Figures

References
-
- Lill NL, Douillard P, Awwad RA, Ota S, Lupher ML, Jr., Miyake S, Meissner-Lula N, Hsu VW, Band H. The evolutionarily conserved N-terminal region of Cbl is sufficient to enhance down-regulation of the epidermal growth factor receptor. J. Biol. Chem. 2000;275:367–377. - PubMed
-
- Duan L, Miura Y, Dimri M, Majumder B, Dodge IL, Reddi AL, Ghosh A, Fernandes N, Zhou P, Mullane-Robinson K, Rao N, Donoghue S, Rogers RA, Bowtell D, Naramura M, Gu H, Band V, Band H. Cbl-mediated ubiquitinylation is required for lysosomal sorting of epidermal growth factor receptor but is dispensable for endocytosis. J. Biol. Chem. 2003;278:28950–28960. - PubMed
-
- Visser GD, Lill NL. The Cbl RING finger C-terminal flank controls epidermal growth factor receptor fate downstream of receptor ubiquitination. Exp. Cell Res. 2005;311:281–293. - PubMed
-
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003;5:461–466. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

