Activation and inhibition of histone deacetylase 8 by monovalent cations
- PMID: 20029090
- PMCID: PMC2825397
- DOI: 10.1074/jbc.M109.033399
Activation and inhibition of histone deacetylase 8 by monovalent cations
Abstract
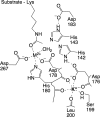
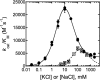
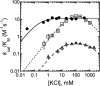
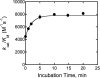
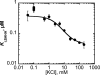
The metal-dependent histone deacetylases (HDACs) catalyze hydrolysis of acetyl groups from acetyllysine side chains and are targets of cancer therapeutics. Two bound monovalent cations (MVCs) of unknown function have been previously observed in crystal structures of HDAC8; site 1 is near the active site, whereas site 2 is located > 20 A from the catalytic metal ion. Here we demonstrate that one bound MVC activates catalytic activity (K(1/2) = 3.4 mM for K(+)), whereas the second, weaker-binding MVC (K(1/2) = 26 mM for K(+)) decreases catalytic activity by 11-fold. The weaker binding MVC also enhances the affinity of the HDAC inhibitor suberoylanilide hydroxamic acid by 5-fold. The site 1 MVC is coordinated by the side chain of Asp-176 that also forms a hydrogen bond with His-142, one of two histidines important for catalytic activity. The D176A and H142A mutants each increase the K(1/2) for potassium inhibition by > or = 40-fold, demonstrating that the inhibitory cation binds to site 1. Furthermore, the MVC inhibition is mediated by His-142, suggesting that this residue is protonated for maximal HDAC8 activity. Therefore, His-142 functions either as an electrostatic catalyst or a general acid. The activating MVC binds in the distal site and causes a time-dependent increase in activity, suggesting that the site 2 MVC stabilizes an active conformation of the enzyme. Sodium binds more weakly to both sites and activates HDAC8 to a lesser extent than potassium. Therefore, it is likely that potassium is the predominant MVC bound to HDAC8 in vivo.
Figures
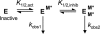
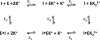
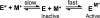
Similar articles
-
Regulation of histone deacetylase 3 by metal cations and 10-hydroxy-2E-decenoic acid: Possible epigenetic mechanisms of queen-worker bee differentiation.PLoS One. 2018 Dec 10;13(12):e0204538. doi: 10.1371/journal.pone.0204538. eCollection 2018. PLoS One. 2018. PMID: 30532259 Free PMC article.
-
Catalytic activity and inhibition of human histone deacetylase 8 is dependent on the identity of the active site metal ion.Biochemistry. 2006 May 16;45(19):6170-8. doi: 10.1021/bi060212u. Biochemistry. 2006. PMID: 16681389
-
General Base-General Acid Catalysis in Human Histone Deacetylase 8.Biochemistry. 2016 Feb 9;55(5):820-32. doi: 10.1021/acs.biochem.5b01327. Epub 2016 Jan 25. Biochemistry. 2016. PMID: 26806311 Free PMC article.
-
Targeting histone deacetylase 8 as a therapeutic approach to cancer and neurodegenerative diseases.Future Med Chem. 2016 Sep;8(13):1609-34. doi: 10.4155/fmc-2016-0117. Epub 2016 Aug 30. Future Med Chem. 2016. PMID: 27572818 Review.
-
Novel structural insights into class I and II histone deacetylases.Curr Top Med Chem. 2009;9(3):235-40. doi: 10.2174/156802609788085304. Curr Top Med Chem. 2009. PMID: 19355988 Review.
Cited by
-
Epigenetic Regulation of TRAIL Signaling: Implication for Cancer Therapy.Cancers (Basel). 2019 Jun 19;11(6):850. doi: 10.3390/cancers11060850. Cancers (Basel). 2019. PMID: 31248188 Free PMC article. Review.
-
Erasers of histone acetylation: the histone deacetylase enzymes.Cold Spring Harb Perspect Biol. 2014 Apr 1;6(4):a018713. doi: 10.1101/cshperspect.a018713. Cold Spring Harb Perspect Biol. 2014. PMID: 24691964 Free PMC article. Review.
-
Inhibition and mechanism of HDAC8 revisited.J Am Chem Soc. 2014 Aug 20;136(33):11636-43. doi: 10.1021/ja501548p. Epub 2014 Aug 7. J Am Chem Soc. 2014. PMID: 25060069 Free PMC article.
-
The structure- and metal-dependent activity of Escherichia coli PgaB provides insight into the partial de-N-acetylation of poly-β-1,6-N-acetyl-D-glucosamine.J Biol Chem. 2012 Sep 7;287(37):31126-37. doi: 10.1074/jbc.M112.390005. Epub 2012 Jul 18. J Biol Chem. 2012. PMID: 22810235 Free PMC article.
-
Discovery of Histone Deacetylase Inhibitor Using Molecular Modeling and Free Energy Calculations.ACS Omega. 2022 May 24;7(22):18786-18794. doi: 10.1021/acsomega.2c01572. eCollection 2022 Jun 7. ACS Omega. 2022. PMID: 35694501 Free PMC article.
References
-
- Glozak M. A., Seto E. (2007) Oncogene 26, 5420–5432 - PubMed
-
- Mann B. S., Johnson J. R., Cohen M. H., Justice R., Pazdur R. (2007) Oncologist 12, 1247–1252 - PubMed
-
- Paris M., Porcelloni M., Binaschi M., Fattori D. (2008) J. Med. Chem. 51, 1505–1529 - PubMed
-
- Gantt S. L., Gattis S. G., Fierke C. A. (2006) Biochemistry 45, 6170–6178 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

