A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease
- PMID: 20029386
- PMCID: PMC2946560
- DOI: 10.1038/tpj.2009.69
A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease
Abstract
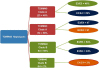
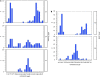
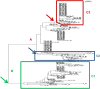
The ɛ4 allele of the apolipoprotein E (APOE) gene is currently the strongest and most highly replicated genetic factor for risk and age of onset of late-onset Alzheimer's disease (LOAD). Using phylogenetic analysis, we have identified a polymorphic poly-T variant, rs10524523, in the translocase of outer mitochondrial membrane 40 homolog (TOMM40) gene that provides greatly increased precision in the estimation of age of LOAD onset for APOE ɛ3 carriers. In two independent clinical cohorts, longer lengths of rs10524523 are associated with a higher risk for LOAD. For APOE ɛ3/4 patients who developed LOAD after 60 years of age, individuals with long poly-T repeats linked to APOE ɛ3 develop LOAD on an average of 7 years earlier than individuals with shorter poly-T repeats linked to APOE ɛ3 (70.5 ± 1.2 years versus 77.6 ± 2.1 years, P=0.02, n=34). Independent mutation events at rs10524523 that occurred during Caucasian evolution have given rise to multiple categories of poly-T length variants at this locus. On replication, these results will have clinical utility for predictive risk estimates for LOAD and for enabling clinical disease prevention studies. In addition, these results show the effective use of a phylogenetic approach for analysis of haplotypes of polymorphisms, including structural polymorphisms, which contribute to complex diseases.
Figures


Similar articles
-
Association and expression analyses with single-nucleotide polymorphisms in TOMM40 in Alzheimer disease.Arch Neurol. 2011 Aug;68(8):1013-9. doi: 10.1001/archneurol.2011.155. Arch Neurol. 2011. PMID: 21825236 Free PMC article.
-
The effect of TOMM40 poly-T length on gray matter volume and cognition in middle-aged persons with APOE ε3/ε3 genotype.Alzheimers Dement. 2011 Jul;7(4):456-65. doi: 10.1016/j.jalz.2010.11.012. Alzheimers Dement. 2011. PMID: 21784354 Free PMC article.
-
TOMM40 rs10524523 polymorphism's role in late-onset Alzheimer's disease and in longevity.J Alzheimers Dis. 2012;28(2):309-22. doi: 10.3233/JAD-2011-110743. J Alzheimers Dis. 2012. PMID: 22008263
-
The effects of the TOMM40 poly-T alleles on Alzheimer's disease phenotypes.Alzheimers Dement. 2018 May;14(5):692-698. doi: 10.1016/j.jalz.2018.01.015. Epub 2018 Mar 7. Alzheimers Dement. 2018. PMID: 29524426 Free PMC article. Review.
-
An inherited variable poly-T repeat genotype in TOMM40 in Alzheimer disease.Arch Neurol. 2010 May;67(5):536-41. doi: 10.1001/archneurol.2010.88. Arch Neurol. 2010. PMID: 20457951 Free PMC article. Review.
Cited by
-
Transcript Variants of Genes Involved in Neurodegeneration Are Differentially Regulated by the APOE and MAPT Haplotypes.Genes (Basel). 2021 Mar 15;12(3):423. doi: 10.3390/genes12030423. Genes (Basel). 2021. PMID: 33804213 Free PMC article.
-
Association study of candidate gene polymorphisms with amnestic mild cognitive impairment in a Chinese population.PLoS One. 2012;7(7):e41198. doi: 10.1371/journal.pone.0041198. Epub 2012 Jul 20. PLoS One. 2012. PMID: 22911757 Free PMC article.
-
APOE and neuroenergetics: an emerging paradigm in Alzheimer's disease.Neurobiol Aging. 2013 Apr;34(4):1007-17. doi: 10.1016/j.neurobiolaging.2012.10.011. Epub 2012 Nov 16. Neurobiol Aging. 2013. PMID: 23159550 Free PMC article. Review.
-
A cytosine-thymine (CT)-rich haplotype in intron 4 of SNCA confers risk for Lewy body pathology in Alzheimer's disease and affects SNCA expression.Alzheimers Dement. 2015 Oct;11(10):1133-43. doi: 10.1016/j.jalz.2015.05.011. Epub 2015 Jun 13. Alzheimers Dement. 2015. PMID: 26079410 Free PMC article.
-
The TOMM40 poly-T rs10524523 variant is associated with cognitive performance among non-demented elderly with type 2 diabetes.Eur Neuropsychopharmacol. 2014 Sep;24(9):1492-9. doi: 10.1016/j.euroneuro.2014.06.002. Epub 2014 Jun 13. Eur Neuropsychopharmacol. 2014. PMID: 25044051 Free PMC article.
References
-
- Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer's disease. Alzheimers Dement. 2007;3:186–191. - PubMed
-
- Pericak-Vance MA, Grubber J, Bailey LR, Hedges D, West S, Santoro L, et al. Identification of novel genes in late-onset Alzheimer's disease. Exp Gerontol. 2000;35:1343–1352. - PubMed
-
- Alzheimer's Association Alzheimer's disease facts and figures. Alzheimers Dement. 2008;4:110–133. - PubMed
-
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer's disease. Arch Gen Psychiatry. 2006;63:168–174. - PubMed
-
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

