Leukaemogenesis: more than mutant genes
- PMID: 20029422
- PMCID: PMC2972637
- DOI: 10.1038/nrc2765
Leukaemogenesis: more than mutant genes
Abstract
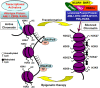
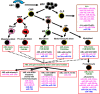
Acute leukaemias are characterized by recurring chromosomal aberrations and gene mutations that are crucial to disease pathogenesis. It is now evident that epigenetic modifications, including DNA methylation and histone modifications, substantially contribute to the phenotype of leukaemia cells. An additional layer of epigenetic complexity is the pathogenetic role of microRNAs in leukaemias, and their key role in the transcriptional regulation of tumour suppressor genes and oncogenes. The genetic heterogeneity of acute leukaemias poses therapeutic challenges, but pharmacological agents that target components of the epigenetic machinery are promising as a component of the therapeutic arsenal for this group of diseases.
Figures
References
-
- Rowley JD. Chromosomal translocations: revisited yet again. Blood. 2008;112:2183–9. - PubMed
-
- Heim S, Mitelman F. Cancer Cytogenetics. Third Edition Wiley-Blackwell; Hoboken, NJ: 2009.
-
- Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

