Reconstructing the initial global spread of a human influenza pandemic: A Bayesian spatial-temporal model for the global spread of H1N1pdm
- PMID: 20029613
- PMCID: PMC2762761
- DOI: 10.1371/currents.RRN1031
Reconstructing the initial global spread of a human influenza pandemic: A Bayesian spatial-temporal model for the global spread of H1N1pdm
Abstract
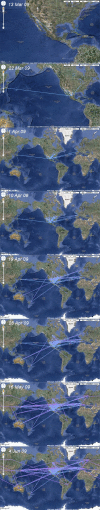
Here, we present an analysis of the H1N1pdm genetic data sampled over the initial stages in the epidemic. To infer phylodynamic spread in time and space we employ a recently developed Bayesian statistical inference framework (Lemey et al., in press). We model spatial diffusion as a continuous-time Markov chain process along time-measured genealogies. In this analysis, we consider 40 locations for which sequence data were available on 06-Aug-2009. The sampling time interval of the 242 sequences spans from 30-Mar-2009 to 12-Jul-2009. The Bayesian inference typically results in a posterior distribution of phylogenetic trees, each having an estimate of the epidemic locations at the ancestral nodes in the tree. We summarize these trees using the most representative clustering pattern and annotate these clusters with the most probable location states. We can visualize this information as tree that grows over time, seeding locations each time an ancestral node is inferred to exist at a different location. A Bayes factor test provides statistical support for epidemiological linkage throughout the evolutionary history. We demonstrate how our full probabilistic approach efficiently tracks an epidemic based on viral genetic data as it unfolds across the globe.
Figures
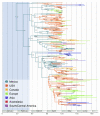
Figure 1. The maximum clade credibility (MCC) tree of H1N1pdm with CTMC spatial reconstruction. Lineages are coloured according to the highest posterior probability for location (these probabilities are shown when > 0.5). The blue band represents the 95% credible interval for the time of the most recent common ancestor.



References
-
- Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team, Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009 Jun 18;360(25):2605-15. Epub 2009 May 7. Erratum in: N Engl J Med. 2009 Jul 2;361(1):102. PubMed PMID: 19423869. - PubMed
-
- Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009 Jun 25;459(7250):1122-5. PubMed PMID: 19516283. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

