Molecular mechanism of elongation factor 1A inhibition by a Legionella pneumophila glycosyltransferase
- PMID: 20030628
- PMCID: PMC3518269
- DOI: 10.1042/BJ20091351
Molecular mechanism of elongation factor 1A inhibition by a Legionella pneumophila glycosyltransferase
Abstract
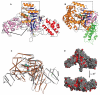
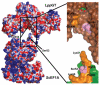
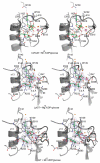
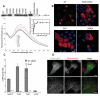
Legionnaires' disease is caused by a lethal colonization of alveolar macrophages with the Gram-negative bacterium Legionella pneumophila. LpGT (L. pneumophila glucosyltransferase; also known as Lgt1) has recently been identified as a virulence factor, shutting down protein synthesis in the human cell by specific glucosylation of EF1A (elongation factor 1A), using an unknown mode of substrate recognition and a retaining mechanism for glycosyl transfer. We have determined the crystal structure of LpGT in complex with substrates, revealing a GT-A fold with two unusual protruding domains. Through structure-guided mutagenesis of LpGT, several residues essential for binding of the UDP-glucose-donor and EF1A-acceptor substrates were identified, which also affected L. pneumophila virulence as demonstrated by microinjection studies. Together, these results suggested that a positively charged EF1A loop binds to a negatively charged conserved groove on the LpGT structure, and that two asparagine residues are essential for catalysis. Furthermore, we showed that two further L. pneumophila glycosyltransferases possessed the conserved UDP-glucose-binding sites and EF1A-binding grooves, and are, like LpGT, translocated into the macrophage through the Icm/Dot (intracellular multiplication/defect in organelle trafficking) system.
Figures

Similar articles
-
Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A.Proc Natl Acad Sci U S A. 2006 Nov 7;103(45):16953-8. doi: 10.1073/pnas.0601562103. Epub 2006 Oct 26. Proc Natl Acad Sci U S A. 2006. PMID: 17068130 Free PMC article.
-
Structural basis of the action of glucosyltransferase Lgt1 from Legionella pneumophila.J Mol Biol. 2010 Feb 19;396(2):321-31. doi: 10.1016/j.jmb.2009.11.044. Epub 2009 Nov 23. J Mol Biol. 2010. PMID: 19941871
-
Region of elongation factor 1A1 involved in substrate recognition by Legionella pneumophila glucosyltransferase Lgt1: identification of Lgt1 as a retaining glucosyltransferase.J Biol Chem. 2009 Jul 24;284(30):20167-74. doi: 10.1074/jbc.M109.008441. Epub 2009 May 28. J Biol Chem. 2009. PMID: 19478083 Free PMC article.
-
Bacterial toxin and effector glycosyltransferases.Biochim Biophys Acta. 2010 Feb;1800(2):134-43. doi: 10.1016/j.bbagen.2009.07.022. Epub 2009 Jul 30. Biochim Biophys Acta. 2010. PMID: 19647041 Review.
-
Cytotoxic glucosyltransferases of Legionella pneumophila.Curr Top Microbiol Immunol. 2013;376:211-26. doi: 10.1007/82_2013_338. Curr Top Microbiol Immunol. 2013. PMID: 23900830 Review.
Cited by
-
Reconstitution of an RNA Virus Replicase in Artificial Giant Unilamellar Vesicles Supports Full Replication and Provides Protection for the Double-Stranded RNA Replication Intermediate.J Virol. 2020 Aug 31;94(18):e00267-20. doi: 10.1128/JVI.00267-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32641477 Free PMC article.
-
Conserved Conformational Hierarchy across Functionally Divergent Glycosyltransferases of the GT-B Structural Superfamily as Determined from Microsecond Molecular Dynamics.Int J Mol Sci. 2021 Apr 28;22(9):4619. doi: 10.3390/ijms22094619. Int J Mol Sci. 2021. PMID: 33924837 Free PMC article.
-
Screening Legionella effectors for antiviral effects reveals Rab1 GTPase as a proviral factor coopted for tombusvirus replication.Proc Natl Acad Sci U S A. 2019 Oct 22;116(43):21739-21747. doi: 10.1073/pnas.1911108116. Epub 2019 Oct 7. Proc Natl Acad Sci U S A. 2019. PMID: 31591191 Free PMC article.
-
VipD of Legionella pneumophila targets activated Rab5 and Rab22 to interfere with endosomal trafficking in macrophages.PLoS Pathog. 2012;8(12):e1003082. doi: 10.1371/journal.ppat.1003082. Epub 2012 Dec 13. PLoS Pathog. 2012. PMID: 23271971 Free PMC article.
-
Targeting Eukaryotic mRNA Translation by Legionella pneumophila.Front Mol Biosci. 2020 Apr 29;7:80. doi: 10.3389/fmolb.2020.00080. eCollection 2020. Front Mol Biosci. 2020. PMID: 32411722 Free PMC article. Review.
References
-
- Coutinho PM, Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert HJ, Davies G, Henrissat B, Svensson B, editors. Recent Advances in Carbohydrate Bioengineering. The Royal Society of Chemistry; Cambridge: 1999. pp. 3–12.
-
- Just I, Selzer J, Wilm M, von Eichel-Streiber C, Mann M, Aktories K. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature. 1995;375:500–503. - PubMed
-
- Schirmer J, Aktories K. Large clostridial cytotoxins: cellular biology of Rho/Ras-glucosylating toxins. Biochim. Biophys. Acta. 2004;1673:66–74. - PubMed
-
- Lyerly D, Wilkins TD. Clostridium difficile. Raven Press; New York: 1995.
-
- Jank T, Giesemann T, Aktories K. Rho-glucosylating Clostridium difficile toxins A and B: new insights into structure and function. Glycobiology. 2007;17:15R–22R. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

