The repertoire of equine intestinal alpha-defensins
- PMID: 20030839
- PMCID: PMC2803202
- DOI: 10.1186/1471-2164-10-631
The repertoire of equine intestinal alpha-defensins
Abstract
Background: Defensins represent an important class of antimicrobial peptides. These effector molecules of the innate immune system act as endogenous antibiotics to protect the organism against infections with pathogenic microorganisms. Mammalian defensins are classified into three distinct sub-families (alpha-, beta- and theta-defensins) according to their specific intramolecular disulfide-bond pattern. The peptides exhibit an antimicrobial activity against a broad spectrum of microorganisms including bacteria and fungi. Alpha-Defensins are primarily synthesised in neutrophils and intestinal Paneth cells. They play a role in the pathogenesis of intestinal diseases and may regulate the flora of the intestinal tract. An equine intestinal alpha-defensin (DEFA1), the first characterised in the Laurasiatheria, shows a broad antimicrobial spectrum against human and equine pathogens. Here we report a first investigation of the repertoire of equine intestinal alpha-defensins. The equine genome was screened for putative alpha-defensin genes by using known alpha-defensin sequences as matrices. Based on the obtained sequence information, a set of oligonucleotides specific to the alpha-defensin gene-family was designed. The products generated by reverse-transcriptase PCR with cDNA from the small intestine as template were sub-cloned and numerous clones were sequenced.
Results: Thirty-eight equine intestinal alpha-defensin transcripts were determined. After translation it became evident that at least 20 of them may code for functional peptides. Ten transcripts lacked matching genomic sequences and for 14 alpha-defensin genes apparently present in the genome no appropriate transcript could be verified. In other cases the same genomic exons were found in different transcripts.
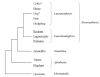
Conclusions: The large repertoire of equine alpha-defensins found in this study points to a particular importance of these peptides regarding animal health and protection from infectious diseases. Moreover, these findings make the horse an excellent species to study biological properties of alpha-defensins. Interestingly, the peptides were not found in other species of the Laurasiatheria to date. Comparison of the obtained transcripts with the genomic sequences in the current assembly of the horse (EquCab2.0) indicates that it is yet not complete and/or to some extent falsely assembled.
Figures


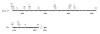
Similar articles
-
A novel horse alpha-defensin: gene transcription, recombinant expression and characterization of the structure and function.Biochem J. 2007 Oct 15;407(2):267-76. doi: 10.1042/BJ20070747. Biochem J. 2007. PMID: 17620056 Free PMC article.
-
Sequence analysis of a 212 kb defensin gene cluster on ECA 27q17.Gene. 2006 Jul 19;376(2):192-8. doi: 10.1016/j.gene.2006.03.006. Epub 2006 Apr 5. Gene. 2006. PMID: 16723195
-
Rapid evolution and diversification of mammalian alpha-defensins as revealed by comparative analysis of rodent and primate genes.Physiol Genomics. 2004 Dec 15;20(1):1-11. doi: 10.1152/physiolgenomics.00150.2004. Epub 2004 Oct 19. Physiol Genomics. 2004. PMID: 15494476
-
Alpha-defensins in the gastrointestinal tract.Mol Immunol. 2003 Nov;40(7):463-7. doi: 10.1016/s0161-5890(03)00157-3. Mol Immunol. 2003. PMID: 14568393 Review.
-
Expression and regulation of antimicrobial peptides in the gastrointestinal tract.J Leukoc Biol. 2004 Jan;75(1):49-58. doi: 10.1189/jlb.0503249. Epub 2003 Oct 2. J Leukoc Biol. 2004. PMID: 14525966 Review.
Cited by
-
Molecular evolution of the primate α-/θ-defensin multigene family.PLoS One. 2014 May 12;9(5):e97425. doi: 10.1371/journal.pone.0097425. eCollection 2014. PLoS One. 2014. PMID: 24819937 Free PMC article.
-
Whole Genome Sequencing Provides New Insights Into the Genetic Diversity and Coat Color of Asiatic Wild Ass and Its Hybrids.Front Genet. 2022 May 12;13:818420. doi: 10.3389/fgene.2022.818420. eCollection 2022. Front Genet. 2022. PMID: 35646088 Free PMC article.
-
An Equine Protein Atlas Highlights Synovial Fluid Proteome Dynamics during Experimentally LPS-Induced Arthritis.J Proteome Res. 2024 Nov 1;23(11):4849-4863. doi: 10.1021/acs.jproteome.4c00125. Epub 2024 Oct 12. J Proteome Res. 2024. PMID: 39395021 Free PMC article.
-
Strain-specific polymorphisms in Paneth cell α-defensins of C57BL/6 mice and evidence of vestigial myeloid α-defensin pseudogenes.Infect Immun. 2011 Jan;79(1):459-73. doi: 10.1128/IAI.00996-10. Epub 2010 Nov 1. Infect Immun. 2011. PMID: 21041494 Free PMC article.
-
Antimicrobial peptides and proteins of the horse--insights into a well-armed organism.Vet Res. 2011 Sep 2;42(1):98. doi: 10.1186/1297-9716-42-98. Vet Res. 2011. PMID: 21888650 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

