Validation of internal reference genes for quantitative real-time PCR in a non-model organism, the yellow-necked mouse, Apodemus flavicollis
- PMID: 20030847
- PMCID: PMC2804578
- DOI: 10.1186/1756-0500-2-264
Validation of internal reference genes for quantitative real-time PCR in a non-model organism, the yellow-necked mouse, Apodemus flavicollis
Abstract
Background: Reference genes are used as internal standards to normalize mRNA abundance in quantitative real-time PCR and thereby allow a direct comparison between samples. So far most of these expression studies used human or classical laboratory model species whereas studies on non-model organism under in-situ conditions are quite rare. However, only studies in free-ranging populations can reveal the effects of natural selection on the expression levels of functional important genes. In order to test the feasibility of gene expression studies in wildlife samples we transferred and validated potential reference genes that were developed for lab mice (Mus musculus) to samples of wild yellow-necked mice, Apodemus flavicollis. The stability and suitability of eight potential reference genes was accessed by the programs BestKeeper, NormFinder and geNorm.
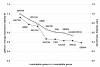
Findings: Although the three programs used different algorithms the ranking order of reference genes was significantly concordant and geNorm differed in only one, NormFinder in two positions compared to BestKeeper. The genes ordered by their mean rank from the most to the least stable gene were: Rps18, Sdha, Canx, Actg1, Pgk1, Ubc, Rpl13a and Actb. Analyses of the normalization factor revealed best results when the five most stable genes were included for normalization.
Discussion: We established a SYBR green qPCR assay for liver samples of wild A. flavicollis and conclude that five genes should be used for appropriate normalization. Our study provides the basis to investigate differential expression of genes under selection under natural selection conditions in liver samples of A. flavicollis. This approach might also be applicable to other non-model organisms.
Figures

References
-
- Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, Bustin SA, Orlando C. Quantitative real-time reverse transcriptionpolymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309(2):293–300. doi: 10.1016/S0003-2697(02)00311-1. - DOI - PubMed
-
- Bas A, Forsberg G, Hammarström S, Hammarström ML. Utility of the Housekeeping Genes 18S rRNA, β-Actin and Glyceraldehyde-3-Phosphate-Dehydrogenase for Normalization in Real-Time Quantitative Reverse Transcriptase-Polymerase Chain Reaction Analysis of Gene Expression in Human T Lymphocytes. Scand J Immunol. 2004;59(6):566–573. doi: 10.1111/j.0300-9475.2004.01440.x. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

