Cardiac resynchronization therapy corrects dyssynchrony-induced regional gene expression changes on a genomic level
- PMID: 20031609
- PMCID: PMC2801868
- DOI: 10.1161/CIRCGENETICS.108.832345
Cardiac resynchronization therapy corrects dyssynchrony-induced regional gene expression changes on a genomic level
Abstract
Background: Cardiac electromechanical dyssynchrony causes regional disparities in workload, oxygen consumption, and myocardial perfusion within the left ventricle. We hypothesized that such dyssynchrony also induces region-specific alterations in the myocardial transcriptome that are corrected by cardiac resynchronization therapy (CRT).
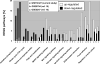
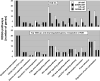
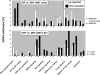
Methods and results: Adult dogs underwent left bundle branch ablation and right atrial pacing at 200 bpm for either 6 weeks (dyssynchronous heart failure, n=12) or 3 weeks, followed by 3 weeks of resynchronization by biventricular pacing at the same pacing rate (CRT, n=10). Control animals without left bundle branch block were not paced (n=13). At 6 weeks, RNA was isolated from the anterior and lateral left ventricular (LV) walls and hybridized onto canine-specific 44K microarrays. Echocardiographically, CRT led to a significant decrease in the dyssynchrony index, while dyssynchronous heart failure and CRT animals had a comparable degree of LV dysfunction. In dyssynchronous heart failure, changes in gene expression were primarily observed in the anterior LV, resulting in increased regional heterogeneity of gene expression within the LV. Dyssynchrony-induced expression changes in 1050 transcripts were reversed by CRT to levels of nonpaced hearts (false discovery rate <5%). CRT remodeled transcripts with metabolic and cell signaling function and greatly reduced regional heterogeneity of gene expression as compared with dyssynchronous heart failure.
Conclusions: Our results demonstrate a profound effect of electromechanical dyssynchrony on the regional cardiac transcriptome, causing gene expression changes primarily in the anterior LV wall. CRT corrected the alterations in gene expression in the anterior wall, supporting a global effect of biventricular pacing on the ventricular transcriptome that extends beyond the pacing site in the lateral wall.
Figures


Comment in
-
Pathways of the heart.Circ Cardiovasc Genet. 2009 Aug;2(4):303-5. doi: 10.1161/CIRCGENETICS.109.892588. Circ Cardiovasc Genet. 2009. PMID: 20031600 Free PMC article. No abstract available.
References
-
- Rosamond W, Flegal K, Furie K, Go A, Greenlund K, Haase N, Hailpern SM, Ho M, Howard V, Kissela B, Kittner S, Lloyd-Jones D, McDermott M, Meigs J, Moy C, Nichol G, O'Donnell C, Roger V, Sorlie P, Steinberger J, Thom T, Wilson M, Hong Y. Heart disease and stroke statistics--2008 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2008;117:e25–146. - PubMed
-
- Baldasseroni S, Opasich C, Gorini M, Lucci D, Marchionni N, Marini M, Campana C, Perini G, Deorsola A, Masotti G, Tavazzi L, Maggioni AP. Left bundle-branch block is associated with increased 1-year sudden and total mortality rate in 5517 outpatients with congestive heart failure: a report from the Italian network on congestive heart failure. Am Heart J. 2002;143:398–405. - PubMed
-
- Bader H, Garrigue S, Lafitte S, Reuter S, Jais P, Haissaguerre M, Bonnet J, Clementy J, Roudaut R. Intra-left ventricular electromechanical asynchrony. A new independent predictor of severe cardiac events in heart failure patients. J Am Coll Cardiol. 2004;43:248–256. - PubMed
-
- Kass DA, Chen CH, Curry C, Talbot M, Berger R, Fetics B, Nevo E. Improved left ventricular mechanics from acute VDD pacing in patients with dilated cardiomyopathy and ventricular conduction delay. Circulation. 1999;99:1567–1573. - PubMed
-
- Spragg DD, Kass DA. Pathobiology of left ventricular dyssynchrony and resynchronization. Prog Cardiovasc Dis. 2006;49:26–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

