Penalized mixtures of factor analyzers with application to clustering high-dimensional microarray data
- PMID: 20031967
- PMCID: PMC2852217
- DOI: 10.1093/bioinformatics/btp707
Penalized mixtures of factor analyzers with application to clustering high-dimensional microarray data
Abstract
Motivation: Model-based clustering has been widely used, e.g. in microarray data analysis. Since for high-dimensional data variable selection is necessary, several penalized model-based clustering methods have been proposed tørealize simultaneous variable selection and clustering. However, the existing methods all assume that the variables are independent with the use of diagonal covariance matrices.
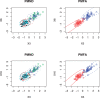
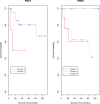
Results: To model non-independence of variables (e.g. correlated gene expressions) while alleviating the problem with the large number of unknown parameters associated with a general non-diagonal covariance matrix, we generalize the mixture of factor analyzers to that with penalization, which, among others, can effectively realize variable selection. We use simulated data and real microarray data to illustrate the utility and advantages of the proposed method over several existing ones.
Figures
Similar articles
-
Mixtures of common t-factor analyzers for clustering high-dimensional microarray data.Bioinformatics. 2011 May 1;27(9):1269-76. doi: 10.1093/bioinformatics/btr112. Epub 2011 Mar 3. Bioinformatics. 2011. PMID: 21372081
-
Variable selection for model-based high-dimensional clustering and its application to microarray data.Biometrics. 2008 Jun;64(2):440-8. doi: 10.1111/j.1541-0420.2007.00922.x. Epub 2007 Oct 26. Biometrics. 2008. PMID: 17970821
-
Penalized model-based clustering with cluster-specific diagonal covariance matrices and grouped variables.Electron J Stat. 2008;2:168-212. doi: 10.1214/08-EJS194. Electron J Stat. 2008. PMID: 19920875 Free PMC article.
-
Computational approaches to analysis of DNA microarray data.Yearb Med Inform. 2006:91-103. Yearb Med Inform. 2006. PMID: 17051302 Review.
-
Comparing algorithms for clustering of expression data: how to assess gene clusters.Methods Mol Biol. 2009;541:479-509. doi: 10.1007/978-1-59745-243-4_21. Methods Mol Biol. 2009. PMID: 19381534 Review.
Cited by
-
Cancer subtype discovery and biomarker identification via a new robust network clustering algorithm.PLoS One. 2013 Jun 17;8(6):e66256. doi: 10.1371/journal.pone.0066256. Print 2013. PLoS One. 2013. PMID: 23799085 Free PMC article.
-
Clustering High-Dimensional Landmark-based Two-dimensional Shape Data‡.J Am Stat Assoc. 2015 Nov 7;110(115):946-961. doi: 10.1080/01621459.2015.1034802. Epub 2015 Apr 16. J Am Stat Assoc. 2015. PMID: 26604425 Free PMC article.
-
Penalized model-based clustering with unconstrained covariance matrices.Electron J Stat. 2009 Jan 1;3:1473-1496. doi: 10.1214/09-EJS487. Electron J Stat. 2009. PMID: 20463857 Free PMC article.
-
Mixture of regressions with multivariate responses for discovering subtypes in Alzheimer's biomarkers with detection limits.Data Sci Sci. 2024;3(1):2309403. doi: 10.1080/26941899.2024.2309403. Epub 2024 Mar 6. Data Sci Sci. 2024. PMID: 38680829 Free PMC article.
References
-
- Baek J, McLachlan GJ. Mixtures of factor analyzers with common factor loadings for the clustering and visualisation of high-dimensional data. Isaac Newton Institute for Mathematical Sciences; 2008. Preprints. - PubMed
-
- Baek J, et al. Mixtures of factor analyzers with common factor loadings: applications to the clustering and visualisation of high-dimensional data. To appear in IEEE Transactions on Pattern Analysis and Machine Intelligence. 2009 Available at: http://www.maths.uq.edu.au/∼gjm/bmf_pami09.pdf/ - PubMed
-
- Beer DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 2002;8:816–824. - PubMed
-
- Dempster AP, et al. Maximum likelihood from incomplete data via the EM algorithm (with discussion) J. R. Stat. Soc. Series B. 1977;39:1–38.

