Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions
- PMID: 20032057
- PMCID: PMC2840681
- DOI: 10.1210/en.2009-0657
Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions
Abstract
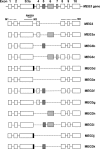
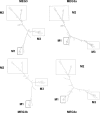
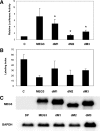
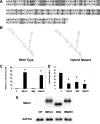
Maternally expressed gene 3 (MEG3) is an imprinted gene highly expressed in the human pituitary. However, MEG3 expression is lost in human gonadotroph-derived pituitary adenomas and most human tumor cell lines. Expression of MEG3 in tumor cells results in growth suppression, p53 protein increase, and activation of p53 downstream targets. The MEG3 gene encodes a noncoding RNA of approximately 1700 nucleotides. There are 12 different MEG3 gene transcripts, generated by alternative splicing. They contain the common exons 1-3 and exons 8-10, but each uses one or more exons 4-7 in a different combination in the middle. MEG3 isoform expression patterns are tissue and cell type specific. Functionally, each isoform stimulates p53-mediated transactivation and suppresses tumor cell growth. We analyzed the secondary RNA folding structure of each MEG3 isoform, using the computer program mfold. All MEG3 RNA isoforms contain three distinct secondary folding motifs M1, M2, and M3. Deletion analysis showed that motifs M2 and M3 are important for p53 activation. Furthermore, a hybrid MEG3 RNA, containing a piece of artificially synthesized sequence different from the wild type but folding into a similar secondary structure, retained the functions of both p53 activation and growth suppression. These results support the hypothesis that a proper folding structure of the MEG3 RNA molecule is critical for its biological functions. This study establishes for the first time the structure-function relationship of a large noncoding RNA and provides a first look into the molecular mechanisms of the biological functions of a large noncoding RNA.
Figures

Similar articles
-
MEG3 noncoding RNA: a tumor suppressor.J Mol Endocrinol. 2012 Apr 2;48(3):R45-53. doi: 10.1530/JME-12-0008. Print 2012 Jun. J Mol Endocrinol. 2012. PMID: 22393162 Free PMC article. Review.
-
Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression.Cancer Res. 2010 Mar 15;70(6):2350-8. doi: 10.1158/0008-5472.CAN-09-3885. Epub 2010 Feb 23. Cancer Res. 2010. PMID: 20179190 Free PMC article.
-
Activation of p53 by MEG3 non-coding RNA.J Biol Chem. 2007 Aug 24;282(34):24731-42. doi: 10.1074/jbc.M702029200. Epub 2007 Jun 13. J Biol Chem. 2007. PMID: 17569660
-
Long Noncoding RNA MEG3 Interacts with p53 Protein and Regulates Partial p53 Target Genes in Hepatoma Cells.PLoS One. 2015 Oct 7;10(10):e0139790. doi: 10.1371/journal.pone.0139790. eCollection 2015. PLoS One. 2015. PMID: 26444285 Free PMC article.
-
Isolation and characterization of novel pituitary tumor related genes: a cDNA representational difference approach.Mol Cell Endocrinol. 2010 Sep 15;326(1-2):40-7. doi: 10.1016/j.mce.2010.02.040. Epub 2010 Mar 6. Mol Cell Endocrinol. 2010. PMID: 20211686 Free PMC article. Review.
Cited by
-
Yeast Telomerase RNA Flexibly Scaffolds Protein Subunits: Results and Repercussions.Molecules. 2020 Jun 14;25(12):2750. doi: 10.3390/molecules25122750. Molecules. 2020. PMID: 32545864 Free PMC article. Review.
-
MEG3 noncoding RNA: a tumor suppressor.J Mol Endocrinol. 2012 Apr 2;48(3):R45-53. doi: 10.1530/JME-12-0008. Print 2012 Jun. J Mol Endocrinol. 2012. PMID: 22393162 Free PMC article. Review.
-
Challenges and perspectives for structural biology of lncRNAs-the example of the Xist lncRNA A-repeats.J Mol Cell Biol. 2019 Oct 25;11(10):845-859. doi: 10.1093/jmcb/mjz086. J Mol Cell Biol. 2019. PMID: 31336384 Free PMC article.
-
The Long Noncoding RNA MEG3 and its Target miR-147 Regulate JAK/STAT Pathway in Advanced Chronic Myeloid Leukemia.EBioMedicine. 2018 Aug;34:61-75. doi: 10.1016/j.ebiom.2018.07.013. Epub 2018 Jul 31. EBioMedicine. 2018. PMID: 30072211 Free PMC article.
-
Novel genetic variants in long non-coding RNA MEG3 are associated with the risk of asthma.PeerJ. 2023 Jan 27;11:e14760. doi: 10.7717/peerj.14760. eCollection 2023. PeerJ. 2023. PMID: 36726728 Free PMC article.
References
-
- Schuster-Gossler K, Simon-Chazottes D, Guenet JL, Zachgo J, Gossler A 1996 Gtl2lacZ, an insertional mutation on mouse chromosome 12 with parental origin-dependent phenotype. Mamm Genome 7:20–24 - PubMed
-
- Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F 2000 Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 5:211–220 - PubMed
-
- Schuster-Gossler K, Bilinski P, Sado T, Ferguson-Smith A, Gossler A 1998 The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev Dyn 212:214–228 - PubMed
-
- Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A 2003 A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab 88:5119–5126 - PubMed
-
- Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A 2007 Activation of p53 by MEG3 non-coding RNA. J Biol Chem 282:24731–24742 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

