Identification of the CD163 protein domains involved in infection of the porcine reproductive and respiratory syndrome virus
- PMID: 20032174
- PMCID: PMC2826032
- DOI: 10.1128/JVI.02093-09
Identification of the CD163 protein domains involved in infection of the porcine reproductive and respiratory syndrome virus
Abstract
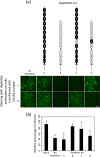
Scavenger receptor CD163 is a key entry mediator for porcine reproductive and respiratory syndrome virus (PRRSV). To identify the CD163 protein domains involved in PRRSV infection, deletion mutants and chimeric mutants were created. Infection experiments revealed that scavenger receptor cysteine-rich (SRCR) domain 5 (SRCR 5) is essential for PRRSV infection, while the four N-terminal SRCR domains and the cytoplasmic tail are not required. The remaining CD163 protein domains need to be present but can be replaced by corresponding SRCR domains from CD163-L1, resulting in reduced (SRCR 6 and interdomain regions) or unchanged (SRCR 7 to SRCR 9) infection efficiency. In addition, CD163-specific antibodies recognizing SRCR 5 are able to reduce PRRSV infection.
Figures

Similar articles
-
The Crystal Structure of the Fifth Scavenger Receptor Cysteine-Rich Domain of Porcine CD163 Reveals an Important Residue Involved in Porcine Reproductive and Respiratory Syndrome Virus Infection.J Virol. 2017 Jan 18;91(3):e01897-16. doi: 10.1128/JVI.01897-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881657 Free PMC article.
-
Replacement of Porcine CD163 Scavenger Receptor Cysteine-Rich Domain 5 with a CD163-Like Homolog Confers Resistance of Pigs to Genotype 1 but Not Genotype 2 Porcine Reproductive and Respiratory Syndrome Virus.J Virol. 2017 Jan 3;91(2):e01521-16. doi: 10.1128/JVI.01521-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27847356 Free PMC article.
-
Deletion of the cytoplasmic domain of CD163 enhances porcine reproductive and respiratory syndrome virus replication.Arch Virol. 2010 Aug;155(8):1319-23. doi: 10.1007/s00705-010-0699-8. Epub 2010 May 23. Arch Virol. 2010. PMID: 20496088
-
A brief review of CD163 and its role in PRRSV infection.Virus Res. 2010 Dec;154(1-2):98-103. doi: 10.1016/j.virusres.2010.07.018. Epub 2010 Jul 23. Virus Res. 2010. PMID: 20655964 Review.
-
Role of CD163 in PRRSV infection.Virology. 2024 Dec;600:110262. doi: 10.1016/j.virol.2024.110262. Epub 2024 Oct 16. Virology. 2024. PMID: 39423600 Review.
Cited by
-
Monkey Viperin Restricts Porcine Reproductive and Respiratory Syndrome Virus Replication.PLoS One. 2016 May 27;11(5):e0156513. doi: 10.1371/journal.pone.0156513. eCollection 2016. PLoS One. 2016. PMID: 27232627 Free PMC article.
-
Antiviral Strategies of Chinese Herbal Medicine Against PRRSV Infection.Front Microbiol. 2020 Jul 28;11:1756. doi: 10.3389/fmicb.2020.01756. eCollection 2020. Front Microbiol. 2020. PMID: 32849384 Free PMC article. Review.
-
Direct Interaction Between CD163 N-Terminal Domain and MYH9 C-Terminal Domain Contributes to Porcine Reproductive and Respiratory Syndrome Virus Internalization by Permissive Cells.Front Microbiol. 2019 Aug 6;10:1815. doi: 10.3389/fmicb.2019.01815. eCollection 2019. Front Microbiol. 2019. PMID: 31447818 Free PMC article.
-
MYH9 Key Amino Acid Residues Identified by the Anti-Idiotypic Antibody to Porcine Reproductive and Respiratory Syndrome Virus Glycoprotein 5 Involve in the Virus Internalization by Porcine Alveolar Macrophages.Viruses. 2019 Dec 29;12(1):40. doi: 10.3390/v12010040. Viruses. 2019. PMID: 31905776 Free PMC article.
-
CD163 and pAPN double-knockout pigs are resistant to PRRSV and TGEV and exhibit decreased susceptibility to PDCoV while maintaining normal production performance.Elife. 2020 Sep 2;9:e57132. doi: 10.7554/eLife.57132. Elife. 2020. PMID: 32876563 Free PMC article.
References
-
- An, T.-Q., Z.-J. Tian, Y.-X. He, Y. Xiao, Y.-F. Jiang, J.-M. Peng, Y.-J. Zhou, D. Liu, and G.-Z. Tong. 5 December 2009, posting date. Porcine reproductive and respiratory syndrome virus attachment is mediated by the N-terminal domain of the sialoadhesin receptor. Vet. Microbiol. doi: 10.1016/j.vetmic.2009.11.006. - DOI - PubMed
-
- Delputte, P. L., W. Van Breedam, I. Delrue, C. Oetke, P. R. Crocker, and H. J. Nauwynck. 2007. Porcine arterivirus attachment to the macrophage-specific receptor sialoadhesin is dependent on the sialic acid-binding activity of the N-terminal immunoglobulin domain of sialoadhesin. J. Virol. 81:9546-9550. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

