The structure of the poxvirus A33 protein reveals a dimer of unique C-type lectin-like domains
- PMID: 20032175
- PMCID: PMC2820914
- DOI: 10.1128/JVI.02247-09
The structure of the poxvirus A33 protein reveals a dimer of unique C-type lectin-like domains
Abstract
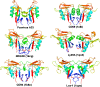
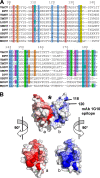
The current vaccine against smallpox is an infectious form of vaccinia virus that has significant side effects. Alternative vaccine approaches using recombinant viral proteins are being developed. A target of subunit vaccine strategies is the poxvirus protein A33, a conserved protein in the Chordopoxvirinae subfamily of Poxviridae that is expressed on the outer viral envelope. Here we have determined the structure of the A33 ectodomain of vaccinia virus. The structure revealed C-type lectin-like domains (CTLDs) that occur as dimers in A33 crystals with five different crystal lattices. Comparison of the A33 dimer models shows that the A33 monomers have a degree of flexibility in position within the dimer. Structural comparisons show that the A33 monomer is a close match to the Link module class of CTLDs but that the A33 dimer is most similar to the natural killer (NK)-cell receptor class of CTLDs. Structural data on Link modules and NK-cell receptor-ligand complexes suggest a surface of A33 that could interact with viral or host ligands. The dimer interface is well conserved in all known A33 sequences, indicating an important role for the A33 dimer. The structure indicates how previously described A33 mutations disrupt protein folding and locates the positions of N-linked glycosylations and the epitope of a protective antibody.
Figures



Similar articles
-
Protein A33 responsible for antibody-resistant spread of Vaccinia virus is homologous to C-type lectin-like proteins.Virus Res. 2010 Jul;151(1):97-101. doi: 10.1016/j.virusres.2010.03.004. Epub 2010 Mar 17. Virus Res. 2010. PMID: 20302896
-
Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A.Eur J Immunol. 2015 Jun;45(6):1605-13. doi: 10.1002/eji.201545509. Epub 2015 Apr 28. Eur J Immunol. 2015. PMID: 25826155
-
Structural and Functional Characterization of Anti-A33 Antibodies Reveal a Potent Cross-Species Orthopoxviruses Neutralizer.PLoS Pathog. 2015 Sep 1;11(9):e1005148. doi: 10.1371/journal.ppat.1005148. eCollection 2015 Sep. PLoS Pathog. 2015. PMID: 26325270 Free PMC article.
-
Antibody Recognition of Immunodominant Vaccinia Virus Envelope Proteins.Subcell Biochem. 2017;83:103-126. doi: 10.1007/978-3-319-46503-6_4. Subcell Biochem. 2017. PMID: 28271474 Review.
-
Natural killer cell recognition of HLA class I molecules.Rev Immunogenet. 2000;2(3):433-48. Rev Immunogenet. 2000. PMID: 11256749 Review.
Cited by
-
Epitope mapping by random peptide phage display reveals essential residues for vaccinia extracellular enveloped virion spread.Virol J. 2012 Sep 24;9:217. doi: 10.1186/1743-422X-9-217. Virol J. 2012. PMID: 23006741 Free PMC article.
-
Vaccinia Virus Glycoproteins A33, A34, and B5 Form a Complex for Efficient Endoplasmic Reticulum to trans-Golgi Network Transport.J Virol. 2020 Mar 17;94(7):e02155-19. doi: 10.1128/JVI.02155-19. Print 2020 Mar 17. J Virol. 2020. PMID: 31941777 Free PMC article.
-
Generation of recombinant mAbs to vaccinia virus displaying high affinity and potent neutralization.Microbiol Spectr. 2023 Sep 22;11(5):e0159823. doi: 10.1128/spectrum.01598-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37737634 Free PMC article.
-
Exploring monkeypox virus proteins and rapid detection techniques.Front Cell Infect Microbiol. 2024 May 28;14:1414224. doi: 10.3389/fcimb.2024.1414224. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38863833 Free PMC article. Review.
-
Rational design of a 'two-in-one' immunogen DAM drives potent immune response against mpox virus.Nat Immunol. 2024 Feb;25(2):307-315. doi: 10.1038/s41590-023-01715-7. Epub 2024 Jan 5. Nat Immunol. 2024. PMID: 38182667
References
-
- Appleyard, G., A. J. Hapel, and E. A. Boulter. 1971. An antigenic difference between intracellular and extracellular rabbitpox virus. J. Gen. Virol. 13:9-17. - PubMed
-
- Banerji, S., A. J. Wright, M. Noble, D. J. Mahoney, I. D. Campbell, A. J. Day, and D. G. Jackson. 2007. Structures of the CD44-hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat. Struct. Mol. Biol. 14:234-239. - PubMed
-
- Boyington, J. C., A. N. Riaz, A. Patamawenu, J. E. Coligan, A. G. Brooks, and P. D. Sun. 1999. Structure of CD94 reveals a novel C-type lectin fold: implications for the NK cell-associated CD94/NKG2 receptors. Immunity 10:75-82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

