The latency-associated nuclear antigen interacts with MeCP2 and nucleosomes through separate domains
- PMID: 20032179
- PMCID: PMC2820923
- DOI: 10.1128/JVI.01097-09
The latency-associated nuclear antigen interacts with MeCP2 and nucleosomes through separate domains
Abstract
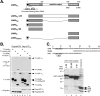
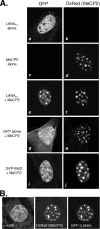
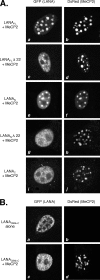
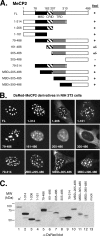
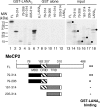
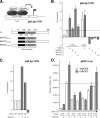
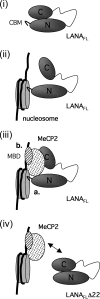
Kaposi's sarcoma-associated herpesvirus (KSHV)-infected cells express the latency-associated nuclear antigen (LANA) involved in the regulation of host and viral gene expression and maintenance of the KSHV latent episome. Performance of these diverse functions involves a 7-amino-acid chromatin-binding motif (CBM) situated at the amino terminus of LANA that is capable of binding directly to nucleosomes. LANA interacts with additional chromatin components, including methyl-CpG-binding protein 2 (MeCP2). Here, we show that the carboxy-terminal DNA-binding/dimerization domain of LANA provides the principal interaction with MeCP2 but that this association is modulated by the CBM. Both domains are required for LANA to colocalize with MeCP2 at chromocenters, regions of extensive pericentric heterochromatin that can be imaged by fluorescence microscopy. Within MeCP2, the methyl-CpG-binding domain (MBD) is the primary determinant for chromatin localization and acts together with the adjacent repression domains (the transcription repression domain [TRD] and the corepressor-interacting domain [CRID]) to redirect LANA to chromocenters. MeCP2 facilitates repression by LANA bound to the KSHV terminal repeats, a function that requires the MeCP2 C terminus in addition to the MBD and CRID/TRD. LANA and MeCP2 can also cooperate to stimulate transcription of the human E2F1 promoter, which lacks a LANA DNA-binding sequence, but this function requires both the N and C termini of LANA. The ability of LANA to establish multivalent interactions with histones and chromatin-binding proteins such as MeCP2 would enable LANA to direct regulatory complexes to specific chromosomal sites and thereby achieve stable reprogramming of cellular gene expression in latently infected cells.
Figures
Similar articles
-
Unidirectional recruitment between MeCP2 and KSHV-encoded LANA revealed by CRISPR/Cas9 recruitment assay.PLoS Pathog. 2025 Mar 10;21(3):e1012972. doi: 10.1371/journal.ppat.1012972. eCollection 2025 Mar. PLoS Pathog. 2025. PMID: 40063648 Free PMC article.
-
Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes.J Virol. 2002 Nov;76(22):11596-604. doi: 10.1128/jvi.76.22.11596-11604.2002. J Virol. 2002. PMID: 12388720 Free PMC article.
-
Site-specific association with host and viral chromatin by Kaposi's sarcoma-associated herpesvirus LANA and its reversal during lytic reactivation.J Virol. 2014 Jun;88(12):6762-77. doi: 10.1128/JVI.00268-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696474 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen: Replicating and Shielding Viral DNA during Viral Persistence.J Virol. 2017 Jun 26;91(14):e01083-16. doi: 10.1128/JVI.01083-16. Print 2017 Jul 15. J Virol. 2017. PMID: 28446671 Free PMC article. Review.
-
The latency-associated nuclear antigen, a multifunctional protein central to Kaposi's sarcoma-associated herpesvirus latency.Future Microbiol. 2011 Dec;6(12):1399-413. doi: 10.2217/fmb.11.137. Future Microbiol. 2011. PMID: 22122438 Free PMC article. Review.
Cited by
-
Latently KSHV-Infected Cells Promote Further Establishment of Latency upon Superinfection with KSHV.Int J Mol Sci. 2021 Nov 5;22(21):11994. doi: 10.3390/ijms222111994. Int J Mol Sci. 2021. PMID: 34769420 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus-encoded LANA recruits topoisomerase IIβ for latent DNA replication of the terminal repeats.J Virol. 2012 Sep;86(18):9983-94. doi: 10.1128/JVI.00839-12. Epub 2012 Jul 3. J Virol. 2012. PMID: 22761383 Free PMC article.
-
The chromatin landscape of Kaposi's sarcoma-associated herpesvirus.Viruses. 2013 May 23;5(5):1346-73. doi: 10.3390/v5051346. Viruses. 2013. PMID: 23698402 Free PMC article. Review.
-
MeCP2 and Chromatin Compartmentalization.Cells. 2020 Apr 3;9(4):878. doi: 10.3390/cells9040878. Cells. 2020. PMID: 32260176 Free PMC article. Review.
-
Epigenetic reprogramming of host genes in viral and microbial pathogenesis.Trends Microbiol. 2010 Oct;18(10):439-47. doi: 10.1016/j.tim.2010.07.003. Epub 2010 Aug 18. Trends Microbiol. 2010. PMID: 20724161 Free PMC article. Review.
References
-
- Amir, R. E., I. B. Van den Veyver, M. Wan, C. Q. Tran, U. Francke, and H. Y. Zoghbi. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23:185-188. - PubMed
-
- An, F. Q., N. Compitello, E. Horwitz, M. Sramkoski, E. S. Knudsen, and R. Renne. 2005. The latency-associated nuclear antigen of Kaposi's sarcoma-asociated herpesvirus modulates cellular gene expression and protects lymphoid cells from P16INK4A-induced cell cycle arrest. J. Biol. Chem. 280:3862-3874. - PubMed
-
- An, J., Y. Sun, and M. B. Rettig. 2004. Transcriptional coactivation of c-Jun by the KSHV-encoded LANA. Blood 103:222-228. - PubMed
-
- Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

