Viral sanctuaries during highly active antiretroviral therapy in a nonhuman primate model for AIDS
- PMID: 20032180
- PMCID: PMC2826073
- DOI: 10.1128/JVI.02356-09
Viral sanctuaries during highly active antiretroviral therapy in a nonhuman primate model for AIDS
Abstract
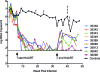
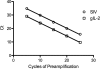
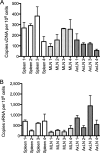
Highly active antiretroviral therapy (HAART) enables long-term suppression of plasma HIV-1 loads in infected persons, but low-level virus persists and rebounds following cessation of therapy. During HAART, this virus resides in latently infected cells, such as resting CD4(+) T cells, and in other cell types that may support residual virus replication. Therapeutic eradication will require elimination of virus from all reservoirs. We report here a comprehensive analysis of these reservoirs in fluids, cells, and tissues in a rhesus macaque model that mimics HAART in HIV-infected humans. This nonhuman primate model uses RT-SHIV, a chimera of simian immunodeficiency virus containing the HIV-1 reverse transcriptase (RT). Methods were developed for extraction, preamplification, and real-time PCR analyses of viral DNA (vDNA) and viral RNA (vRNA) in tissues from RT-SHIV-infected macaques. These methods were used to identify viral reservoirs in RT-SHIV-infected macaques treated with a potent HAART regimen consisting of efavirenz, emtricitabine, and tenofovir. Plasma virus loads at necropsy ranged from 11 to 28 copies of vRNA per ml. Viral RNA and DNA were detected during HAART, in tissues from numerous anatomical locations. Additional analysis provided evidence for full-length viral RNA in tissues of animals with virus suppressed by HAART. The highest levels of vDNA and vRNA in HAART-treated macaques were in lymphoid tissues, particularly the spleen, lymph nodes, and gastrointestinal tract tissues. This study is the first comprehensive analysis of the tissue and organ distribution of a primate AIDS virus during HAART. These data demonstrate widespread persistence of residual virus in tissues during HAART.
Figures

References
-
- Adams, M., L. Sharmeen, J. Kimpton, J. M. Romeo, J. V. Garcia, B. M. Peterlin, M. Groudine, and M. Emerman. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. USA 91:3862-3866. - PMC - PubMed
-
- Ambrose, Z., V. Boltz, S. Palmer, J. M. Coffin, S. H. Hughes, and V. N. Kewalramani. 2004. In vitro characterization of a simian immunodeficiency virus-human immunodeficiency virus (HIV) chimera expressing HIV type 1 reverse transcriptase to study antiviral resistance in pigtail macaques. J. Virol. 78:13553-13561. - PMC - PubMed
-
- Ambrose, Z., S. Palmer, V. F. Boltz, M. Kearney, K. Larsen, P. Polacino, L. Flanary, K. Oswald, M. Piatak, Jr., J. Smedley, W. Shao, N. Bischofberger, F. Maldarelli, J. T. Kimata, J. W. Mellors, S. L. Hu, J. M. Coffin, J. D. Lifson, and V. N. KewalRamani. 2007. Suppression of viremia and evolution of human immunodeficiency virus type 1 drug resistance in a macaque model for antiretroviral therapy. J. Virol. 81:12145-12155. - PMC - PubMed
-
- Bailey, J. R., A. R. Sedaghat, T. Kieffer, T. Brennan, P. K. Lee, M. Wind-Rotolo, C. M. Haggerty, A. R. Kamireddi, Y. Liu, J. Lee, D. Persaud, J. E. Gallant, J. Cofrancesco, Jr., T. C. Quinn, C. O. Wilke, S. C. Ray, J. D. Siliciano, R. E. Nettles, and R. F. Siliciano. 2006. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 80:6441-6457. - PMC - PubMed
-
- Balzarini, J., E. De Clercq, and K. Uberla. 1997. SIV/HIV-1 hybrid virus expressing the reverse transcriptase gene of HIV-1 remains sensitive to HIV-1-specific reverse transcriptase inhibitors after passage in rhesus macaques. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 15:1-4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

