The foamy virus genome remains unintegrated in the nuclei of G1/S phase-arrested cells, and integrase is critical for preintegration complex transport into the nucleus
- PMID: 20032182
- PMCID: PMC2826029
- DOI: 10.1128/JVI.02435-09
The foamy virus genome remains unintegrated in the nuclei of G1/S phase-arrested cells, and integrase is critical for preintegration complex transport into the nucleus
Abstract
Foamy viruses are a member of the spumavirus subfamily of retroviruses with unique mechanisms of virus replication. Foamy virus replication is cell cycle dependent; however, the genome is found in the nuclei of cells arrested in the G(1)/S phase. Despite the presence of genome in the nuclei of growth-arrested cells, there is no viral gene expression, thus explaining its dependency on cell cycle. This report shows that the foamy virus genome remains unintegrated in G(1)/S phase-arrested cells. The foamy virus genome is detected by confocal microscopy in the nuclei of both dividing and growth-arrested cells. Alu PCR revealed foamy virus-specific DNA amplification from genomic DNA isolated in cycling cells at 24 h postinfection. In arrested cells no foamy virus DNA band was detected in cells harvested at 1 or 7 days after infection, and a very faint band that is significantly less than DNA amplified from cycling cells was observed at day 15. After these cells were arrested at the G(1)/S phase for 1, 7, or 15 days they were allowed to cycle, at which time foamy virus-specific DNA amplification was readily observed. Taken together, these results suggest that the foamy virus genome persists in nondividing cells without integrating. We have also established evidence for the first time that the foamy virus genome and Gag translocation into the nucleus are dependent on integrase in cycling cells, implicating the role of integrase in transport of the preintegration complex into the nucleus. Furthermore, despite the presence of a nuclear localization signal sequence in Gag, we observed no foamy virus Gag importation into the nucleus in the absence of integrase.
Figures


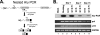
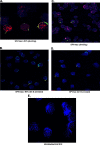
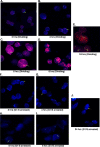

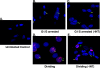
Similar articles
-
Foamy Virus Integrase in Development of Viral Vector for Gene Therapy.J Microbiol Biotechnol. 2020 Sep 28;30(9):1273-1281. doi: 10.4014/jmb.2003.03046. J Microbiol Biotechnol. 2020. PMID: 32699199 Free PMC article. Review.
-
Integrase C-terminal residues determine the efficiency of feline foamy viral DNA integration.Virology. 2018 Jan 15;514:50-56. doi: 10.1016/j.virol.2017.10.022. Epub 2017 Nov 10. Virology. 2018. PMID: 29128756
-
Nuclear localization signals in prototype foamy viral integrase for successive infection and replication in dividing cells.Mol Cells. 2014 Feb;37(2):140-8. doi: 10.14348/molcells.2014.2331. Epub 2014 Feb 19. Mol Cells. 2014. PMID: 24598999 Free PMC article.
-
An active foamy virus integrase is required for virus replication.J Gen Virol. 1999 Jun;80 ( Pt 6):1445-1452. doi: 10.1099/0022-1317-80-6-1445. J Gen Virol. 1999. PMID: 10374962
-
The replication strategy of foamy viruses.Curr Top Microbiol Immunol. 2003;277:1-26. doi: 10.1007/978-3-642-55701-9_1. Curr Top Microbiol Immunol. 2003. PMID: 12908766 Review.
Cited by
-
Molecular biology of foamy viruses.Med Microbiol Immunol. 2010 Aug;199(3):197-207. doi: 10.1007/s00430-010-0158-x. Epub 2010 May 6. Med Microbiol Immunol. 2010. PMID: 20445989 Review.
-
Foamy virus biology and its application for vector development.Viruses. 2011 May;3(5):561-85. doi: 10.3390/v3050561. Epub 2011 May 11. Viruses. 2011. PMID: 21994746 Free PMC article. Review.
-
Prototype foamy virus gag nuclear localization: a novel pathway among retroviruses.J Virol. 2011 Sep;85(18):9276-85. doi: 10.1128/JVI.00663-11. Epub 2011 Jun 29. J Virol. 2011. PMID: 21715475 Free PMC article.
-
Foamy Virus Integrase in Development of Viral Vector for Gene Therapy.J Microbiol Biotechnol. 2020 Sep 28;30(9):1273-1281. doi: 10.4014/jmb.2003.03046. J Microbiol Biotechnol. 2020. PMID: 32699199 Free PMC article. Review.
-
Structural and functional insights into foamy viral integrase.Viruses. 2013 Jul 18;5(7):1850-66. doi: 10.3390/v5071850. Viruses. 2013. PMID: 23872492 Free PMC article. Review.
References
-
- An, D. G., U. Hyun, and C. G. Shin. 2008. Characterization of nuclear localization signals of the prototype foamy virus integrase. J. Gen. Virol. 89:1680-1684. - PubMed
-
- Bahner, I., K. Kearns, Q. L. Hao, E. M. Smogorzewska, and D. B. Kohn. 1996. Transduction of human CD34+ hematopoietic progenitor cells by a retroviral vector expressing an RRE decoy inhibits human immunodeficiency virus type 1 replication in myelomonocytic cells produced in long-term culture. J. Virol. 70:4352-4360. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

