The latent origin of replication of Epstein-Barr virus directs viral genomes to active regions of the nucleus
- PMID: 20032186
- PMCID: PMC2820910
- DOI: 10.1128/JVI.01909-09
The latent origin of replication of Epstein-Barr virus directs viral genomes to active regions of the nucleus
Abstract
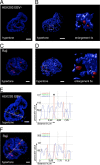
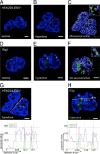
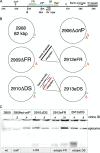
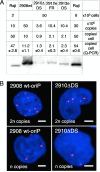
The Epstein-Barr virus efficiently infects human B cells. The EBV genome is maintained extrachromosomally and replicates synchronously with the host's chromosomes. The latent origin of replication (oriP) guarantees plasmid stability by mediating two basic functions: replication and segregation of the viral genome. While the segregation process of EBV genomes is well understood, little is known about its chromatin association and nuclear distribution during interphase. Here, we analyzed the nuclear localization of EBV genomes and the role of functional oriP domains FR and DS for basic functions such as the transformation of primary cells, their role in targeting EBV genomes to distinct nuclear regions, and their association with epigenetic domains. Fluorescence in situ hybridization visualized the localization of extrachromosomal EBV genomes in the regions adjacent to chromatin-dense territories called the perichromatin. Further, immunofluorescence experiments demonstrated a preference of the viral genome for histone 3 lysine 4-trimethylated (H3K4me3) and histone 3 lysine 9-acetylated (H3K9ac) nuclear regions. To determine the role of FR and DS for establishment and subnuclear localization of EBV genomes, we transformed primary human B lymphocytes with recombinant mini-EBV genomes containing different oriP mutants. The loss of DS results in a slightly increased association in H3K27me3 domains. This study demonstrates that EBV genomes or oriP-based extrachromosomal vector systems are integrated into the higher order nuclear organization. We found that viral genomes are not randomly distributed in the nucleus. FR but not DS is crucial for the localization of EBV in perichromatic regions that are enriched for H3K4me3 and H3K9ac, which are hallmarks of transcriptionally active regions.
Figures


Similar articles
-
Replication licensing of the EBV oriP minichromosome.Curr Top Microbiol Immunol. 2001;258:13-33. doi: 10.1007/978-3-642-56515-1_2. Curr Top Microbiol Immunol. 2001. PMID: 11443858 Review.
-
Epstein-Barr Virus Episome Physically Interacts with Active Regions of the Host Genome in Lymphoblastoid Cells.J Virol. 2020 Nov 23;94(24):e01390-20. doi: 10.1128/JVI.01390-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32999023 Free PMC article.
-
Initiation of DNA replication within oriP is dispensable for stable replication of the latent Epstein-Barr virus chromosome after infection of established cell lines.J Virol. 2000 Sep;74(18):8563-74. doi: 10.1128/jvi.74.18.8563-8574.2000. J Virol. 2000. PMID: 10954558 Free PMC article.
-
Initiation of latent DNA replication in the Epstein-Barr virus genome can occur at sites other than the genetically defined origin.Mol Cell Biol. 1995 May;15(5):2893-903. doi: 10.1128/MCB.15.5.2893. Mol Cell Biol. 1995. PMID: 7739569 Free PMC article.
-
[Epstein-Barr Virus Genome Replication as a Molecular Target for Cancer Therapy].Yakugaku Zasshi. 2019;139(1):63-67. doi: 10.1248/yakushi.18-00164-1. Yakugaku Zasshi. 2019. PMID: 30606931 Review. Japanese.
Cited by
-
Repair of DNA strand breaks in a minichromosome in vivo: kinetics, modeling, and effects of inhibitors.PLoS One. 2013;8(1):e52966. doi: 10.1371/journal.pone.0052966. Epub 2013 Jan 30. PLoS One. 2013. PMID: 23382828 Free PMC article.
-
The Epstein-Barr Virus EBNA1 Protein.Scientifica (Cairo). 2012;2012:438204. doi: 10.6064/2012/438204. Epub 2012 Dec 19. Scientifica (Cairo). 2012. PMID: 24278697 Free PMC article. Review.
-
The IS2 Element Improves Transcription Efficiency of Integration-Deficient Lentiviral Vector Episomes.Mol Ther Nucleic Acids. 2018 Dec 7;13:16-28. doi: 10.1016/j.omtn.2018.08.007. Epub 2018 Aug 18. Mol Ther Nucleic Acids. 2018. PMID: 30227274 Free PMC article.
-
Changes in gene expression levels caused by H3K9me3/H3K9ac modifications are associated with BmCPV infection in Bombyx mori.Virulence. 2025 Dec;16(1):2510535. doi: 10.1080/21505594.2025.2510535. Epub 2025 May 26. Virulence. 2025. PMID: 40418637 Free PMC article.
-
Evidence for Tethering of Human Cytomegalovirus Genomes to Host Chromosomes.Front Cell Infect Microbiol. 2020 Sep 30;10:577428. doi: 10.3389/fcimb.2020.577428. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33117732 Free PMC article.
References
-
- Albiez, H., M. Cremer, C. Tiberi, L. Vecchio, L. Schermelleh, et al. 2006. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 14:707-733. - PubMed
-
- Bolte, S., and F. P. Cordelieres. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213-232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

